Preparation method of pseudotype virus for nucleic acid detection of 2019-nCoV
A virus nucleic acid, pseudovirus technology, applied in the field of pseudovirus preparation, can solve complex problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1: Construction of a pseudoviral vector that eliminates unnecessary elements of the lentiviral expression vector
[0081] In the existing lentiviral vector FV011 (Fubai’s own vector, such as Picture 8 Shown), remove the promoter and foreign expression box:
[0082] Step S011: Enzyme digestion of the lentiviral vector at 37°C in a water bath for 2 hours. The digestion system is specifically:
[0083]
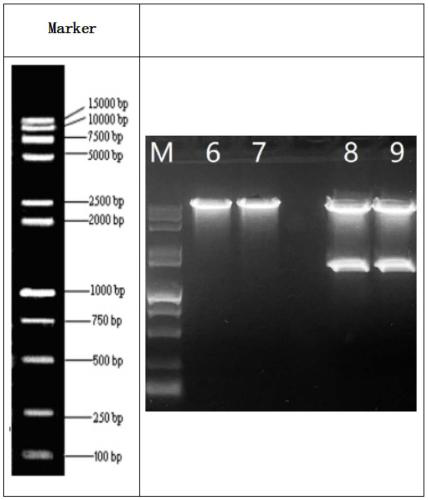
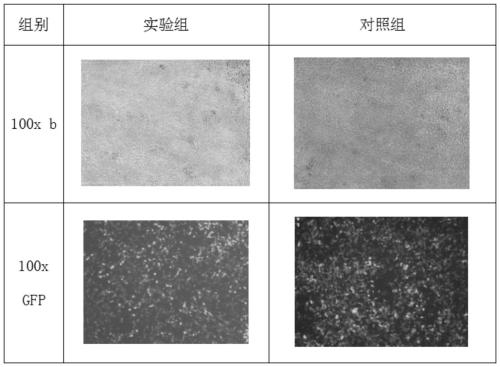
[0084] Step S012: Agarose gel electrophoresis and gel recovery target product: use 1% agarose, 120V constant pressure electrophoresis for 30-40 minutes, cut out a single target fragment under ultraviolet irradiation and put it into a clean centrifuge tube for gel recovery and purification. Loading information: samples 8-9: FV-011-PacI-NheI; the electrophoresis results are as follows figure 1 Shown.
Embodiment 2
[0085] Example 2: Cloning part of the genome of the virus to be detected into a pseudovirus vector;
[0086] Step S021: Obtain the partial sequence of the coronavirus by way of full gene synthesis, and synthesize it into the PUC57 vector, denoted as PUC57-ORF 1a / bEN; the synthetic sequence contains the PAC I restriction site at the 5'end and the Nhe I enzyme at the 3'end The cut site, the specific sequence information is as follows:
[0087] TAATTAAATCGTGTTGTCTGTACTGCCGTTGCCACATAGATCATCCAAATCCTAAAGGATTTTGTGACTTAAAAGGTAAGTATGTACAAATACCTACAACTTGTGCTAATGACCCTGTGGGTTTTACACTTAAAAACACAGTCTGTACCGTCTGCGGTATGTGGAAAGGTTATGGCTGTAGTTGTGATCAACTCCGCGAACCCATGCTTCAGTCAGCTGATGCACAATCGTTTTTAAACGGGTTTGCGGTGTAAGTGCAGCCCGTCTTACACCGTGCGGCACAGGCACTAGTACTGATGTCGTATACAGGGCTTTTGACATCTACAATGATAAAGTAGCTGGTTTTGCTAAATTCCTAAAAACTAATTGTTGTCGCTTCCAAGAAAAGGACGAAGATGACAATTTAATTGATTCTTACTTTGTAGTTAAGAGACACACTTTCTCTAACTACCAACATGAAGAAACAATTTATAATTTACTTAAGGATTGTCCAGCTGTTGCTAAACATATGTACTCATTCGTTTCGGAAGAGACAGGTACGTTAATAGTTA...
Embodiment 3
[0106] Example 3: The integrase gene of the lentivirus packaging helper plasmid is mutated to reduce or lose the integration ability of the lentivirus:
[0107] Before mutation:
[0108] tttttagatggaatagataaggcccaagaagaacatgagaaatatcacagtaattggagagcaatggctagtgattttaacctaccacctgtagtagcaaaagaaatagtagccagctgtgataaatgtcagctaaaaggggaagccatgcatggacaagtagactgtagcccaggaatatggcagcta Gat tgtacacatttagaaggaaaagttatcttggtagcagttcatgtagccagtggatatatagaagcagaagtaattccagcagagacagggcaagaaacagcatacttcctcttaaaattagcaggaagatggccagtaaaaacagtacatacagacaatggcagcaatttcaccagtactacagttaaggccgcctgttggtgggcggggatcaagcaggaatttggcattccctacaatccccaaagtcaaggagtaatagaatctatgaataaagaattaaagaaaattataggacaggtaagagatcaggctgaacatcttaagacagcagtacaaatggcagtattcatccacaattttaaaagaaaaggggggattggggggtacagtgcaggggaaagaatagtagacataatagcaacagacatacaaactaaagaattacaaaaacaaattacaaaaattcaaaattttcgggtttattacagggacagcagagatccagtttggaaaggaccagcaaagctcctctggaaaggtgaaggggcagtagtaatacaagataatagtgacataaaagtagtgcca AGaAGaAaA gcaaagatcatca...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com