SARS-CoV-2 antibody detection method
An antibody detection, sars-cov-2 technology, applied in the field of SARS-CoV-2 antibody detection, can solve the problem of high homology and achieve important application prospects and value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Antigen horseradish peroxidase (HRP) labeling
[0028] (1) Antigen horseradish peroxidase (HRP) labeling:
[0029] (1) Take 6.4 mg of HRP dry powder, add 0.6 ml of double distilled water, slowly add 60 μl of sodium periodate (0.1M) dropwise into HRP, incubate at room temperature for 20 min, and dialyze the sodium acetate solution (1 mM pH4.4) overnight;
[0030] (2) Add 1.6 mg of antigenic protein to the dialyzed HRP solution, and incubate at room temperature for 2 hours;
[0031] The amino acid sequence of the antigenic protein is shown in SEQ ID NO.1, which is prepared by the following method: inserting the human SARS-CoV Nucleocapsid gene (nucleotide sequence shown in SEQ ID NO.2) into the expression vector pET28a, Construct pET28a-SARS-CoV-N fusion expression plasmid, transform into Escherichia coli, induce with IPTG, purify and renature to obtain recombinant SARS-CoV-2 Nucleocapsid protein;
[0032] (3) Add 60 μl of sodium borohydride (4 mg / ml) to the ...
Embodiment 2
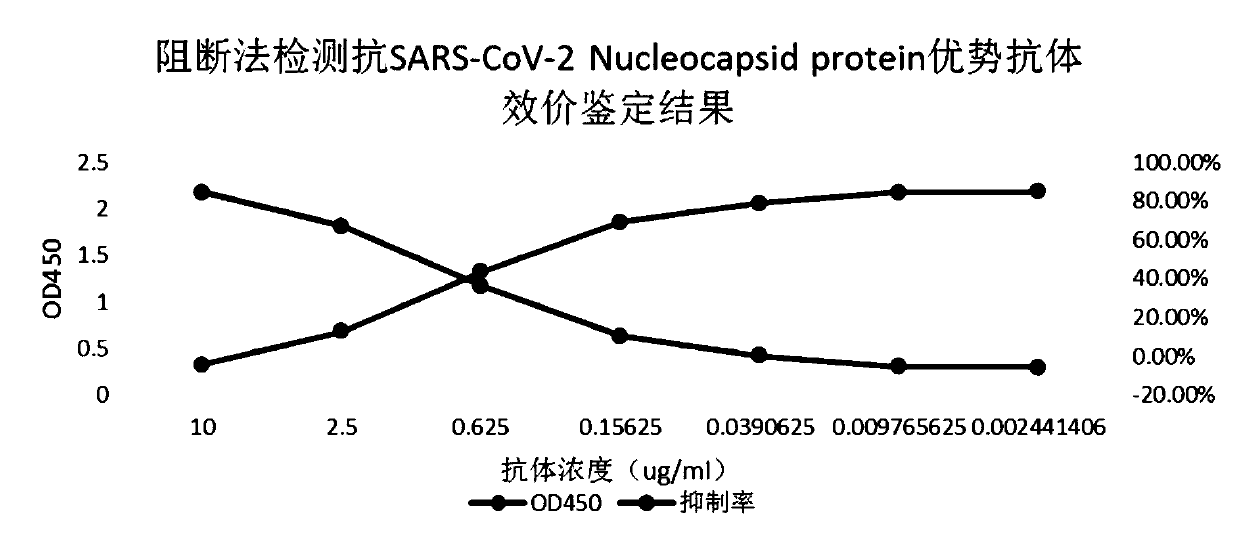
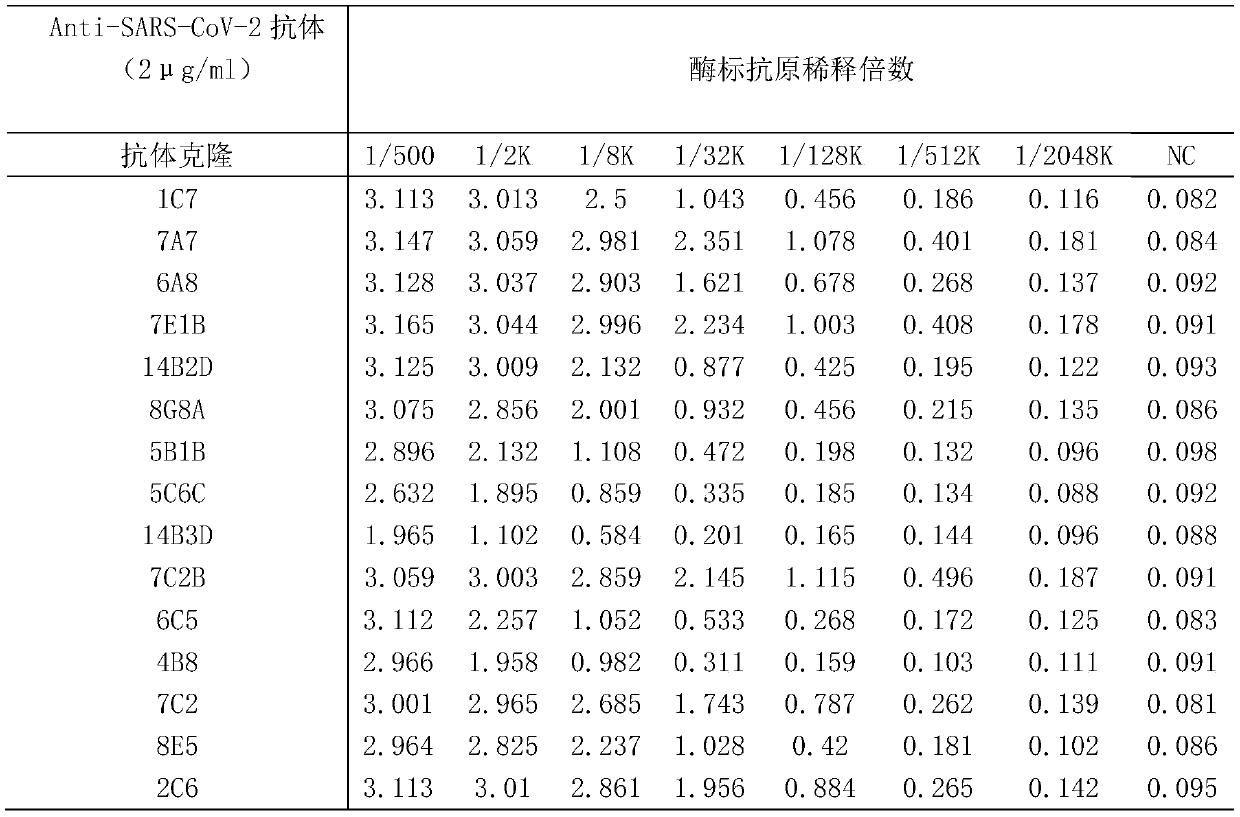
[0044] Embodiment 2: competition experiment screening
[0045] Use checkerboard titration to screen for competition experiments. First, take all Anti-SARS-CoV-2 Nucleocapsid protein-specific antibodies and configure them at a concentration of 2 μg / ml and 1 μg / ml and prepare them into coated plates. The coating and blocking process refer to the examples 1. The experiment was designed according to the checkerboard titration method. The Anti-SARS-CoV-2 Nucleocapsidprotein antibody was prepared into 20ug / ml and 0ug / ml solutions as competing antibodies, and the enzyme-labeled antigen was prepared into 2ug / ml and 1ug / ml concentrations, each of which was 50μl Perform competition detection and incubate at room temperature for 15 minutes. Please refer to Example 1 for the remaining detection, color development, and termination processes. According to the experimental results, select the antibody clone with the highest inhibition rate and the working concentration of the corresponding ...
Embodiment 3
[0049] Embodiment 3: Matrix effect detection of sample
[0050] According to the experimental results obtained in Example 2, select the dominant antibody clone and its relevant raw material working concentration, and further detect the matrix effect of the sample, the specific steps are as follows:
[0051] (1) Serum matrix effect detection
[0052] a. Sample source: Rabbits were immunized with recombinant SARS-CoV-2 Nucleocapsid protein, and the serum after immunization was taken as the positive sample in the experiment.
[0053] b. Coating: 1C7 antibody was prepared to a concentration of 2ug / ml for coating, and other methods were the same as in Example 1.
[0054] Closing: The method is the same as that of implementation case 1.
[0055] Sample dilution: Take normal human serum and dilute it with phosphate buffer saline containing 1% bovine serum albumin, the dilution gradient is shown in Table 3. At the same time, use phosphate buffer saline with 1% bovine serum albumin a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com