RAA primer, probe and detection method for detecting pasteurella multocida diseases
A technology of Pasteurella and multocida, which is applied in the field of bacterial molecular biology detection, can solve the problems of large human subjective factors, complex instruments, and long time-consuming in microscopic examination methods, so as to achieve convenient, fast and accurate identification and reduce detection time , The rapid effect of the detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Select the conserved region sequence of the Pasteurella multocida kmt1 gene to design primers and probes. Find the corresponding full gene sequence in Genebank (www.ncbi.nlm.nih.gov), and use DNASTAR software for homology analysis and Blast Sequence analysis screened out the highly conserved sequence of the Pasteurella multocida kmt1 gene as follows (this fragment is highly conserved in the Pasteurella multocida kmt1 gene, and is compatible with Mycoplasma, Streptococcus, Escherichia coli, Eperythrozoon, Neospore worm, Toxoplasma gondii, healthy bovine blood and other pathogenic gene sequences have no cross-matching reaction and no cross-reaction):
[0038] TATTTTGGCGTGATGAATCAAGCGGTCACAGAAAAGACAGCAATTTCGAGCAAACAATGGTGGGGCTTTACGCTGATTAATATTGTGCTGACATTACTGCTCTATCCGCTATTTACCCAGTGGGGCGGTGCGAATGAACCGATTGCCGCGAAATTGAGTTTTATGCCACTTGAAATGGGAAATGGCATTATTTTATGNO.GCTCGTTGTTGTGAGTGGGCTTG(ID
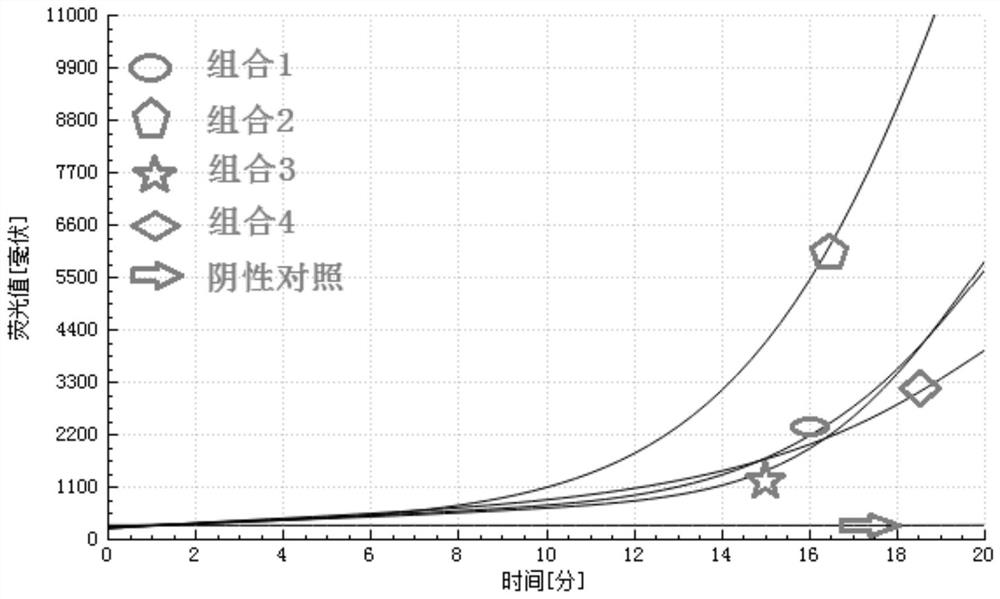
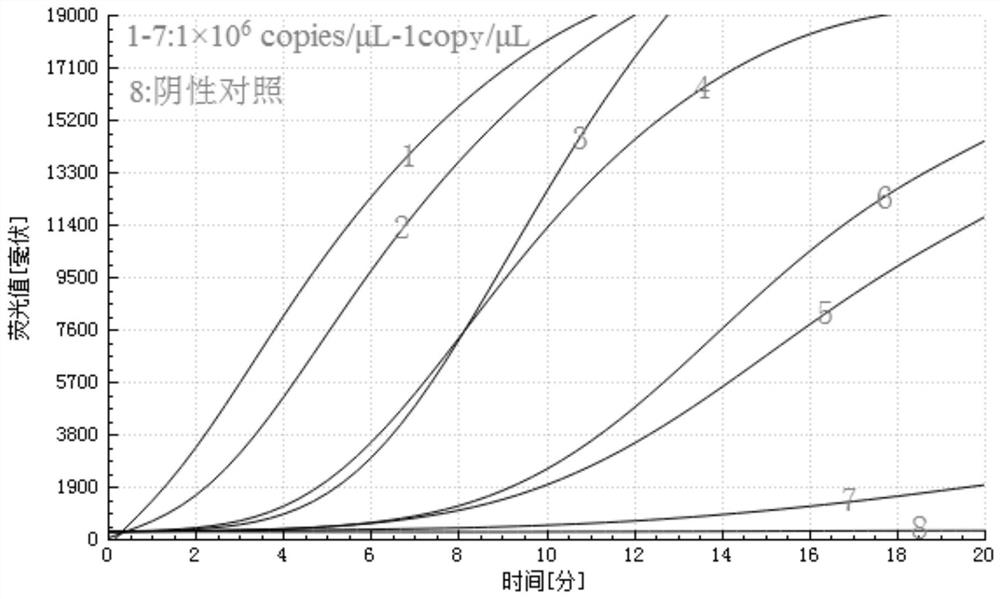
[0039] The highly conserved sequence obtained by screening was used as the target gene fr...
Embodiment 2
[0066] Example 2 The method for detecting Pasteurella multocida by RAA fluorescence method
[0067] The method for detecting Pasteurella multocida by RAA fluorescence method comprises the steps:
[0068] (1) Extract the nucleic acid from the bacterial sample according to the bacterial whole genome method, and store it at -20°C for later use;
[0069] (2) Turn on the constant temperature fluorescent gene detector RAA-F1620 for preheating, and set the reaction parameters. The reaction parameters are set to 39°C, and the reaction time is 20 minutes;
[0070] (3) Add 13.7 μL of water to 25 μL of reaction buffer, 2.1 μL of upstream and downstream primers and 0.6 μL of probe at a concentration of 10 μM, mix well, add to RAA fluorescent basic reaction reagent and mix to obtain a reaction master mix;
[0071] (4) Add 2.5 μL of Mg to the cap of the reaction tube 2+ , fully mixing 4 μL of the nucleic acid extraction solution obtained in the step (1) with the reaction premix solution o...
Embodiment 3
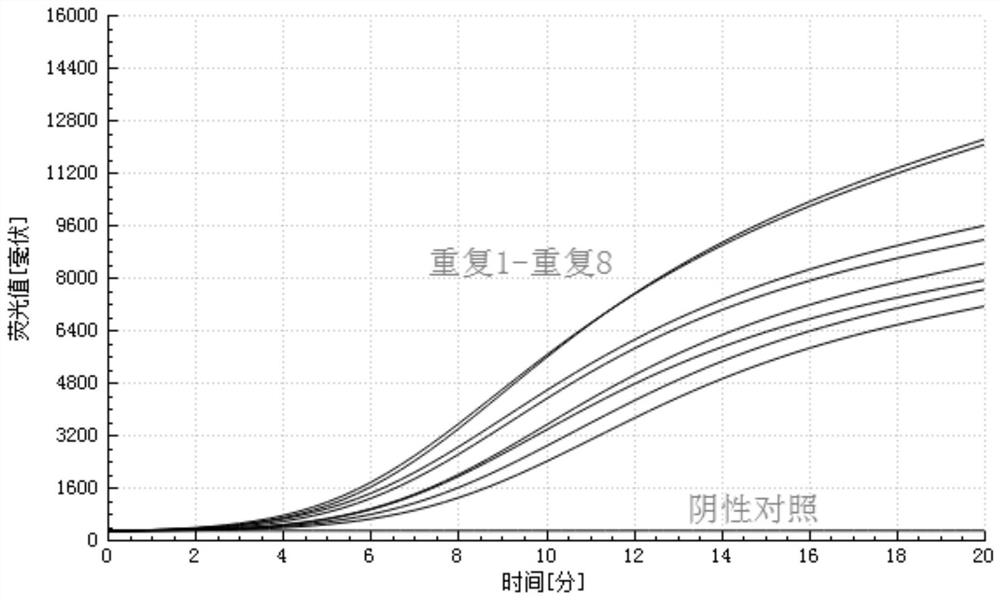
[0137] Embodiment 3 actual sample detection
[0138] (1) The sequences of primers, probes and negative quality controls are the same as in Example 1.
[0139] (2) A total of 15 clinical samples from 1 to 15 in the experiment were provided by Yanbian Prefecture Animal Disease Prevention and Control Center;
[0140] (3) Sample extraction method:
[0141] Extract the nucleic acid from the bacterial liquid sample according to the bacterial genome method, and store it at -20°C for later use;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com