Shewanella for expressing functionalized amyloid fibre, construction method and application thereof
A technology of Shewanella and amyloid, applied in the field of genetic engineering, can solve the problems of no research reports and no research reports, and achieve the effects of rapid reproduction, good adsorption rate, and good commercial application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
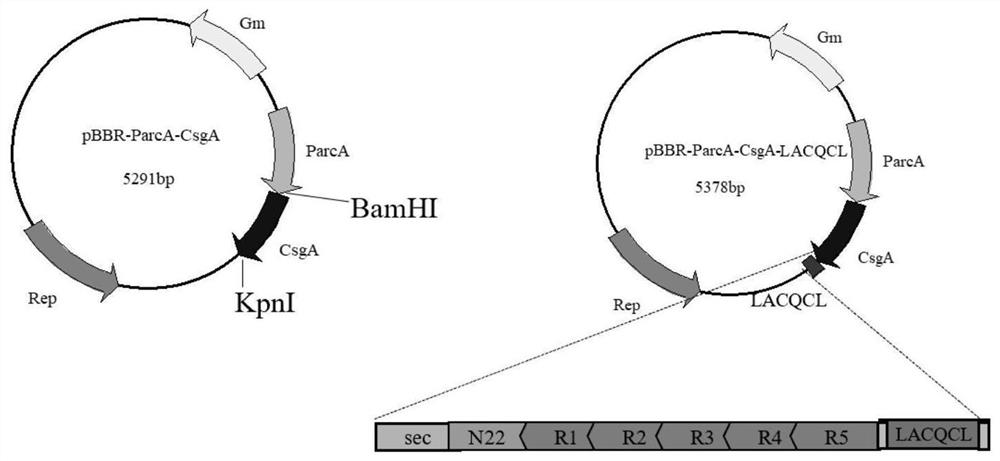
[0028] Example 1: csgA-LACQCL gene synthesis and the construction of recombinant Shewanella expressing functional amyloid fiber curli (csgA-LACQCL)
[0029] Translates the additional peptide domain LACQCL into the nuclear The nucleotide sequence was codon optimized, and the flexible sequence GSGGSG was connected to the C-terminal of the csgA gene to finally construct the gene fragment csgA-LACQCL, whose amino acid sequence is shown in SEQ ID NO.1, and the nucleotide sequence is shown in SEQ ID NO.2 shown.
[0030] Use the Primer Premier 5.0 software to design a primer pair for amplifying the csgA-LACQCL sequence. The forward primer sequence is shown in SEQ ID NO.3, namely: CGCCATATGAAACTTTTAAAAGTAGCAGCAAT, and the reverse primer sequence is two segments, respectively, as in SEQ ID NO.4 As shown, namely: GTGGTGCGCCTGAGCTGTATGCACCTGAACCACCTGAACCGTACTGATGAGCGGTCGCG and as shown in SEQ ID NO.5, namely: CCGCTCGAGTTAACCTGAACCACCTGAACCGAATGGTGGCATTGGTGGTGCGCCTGAGCTG.
[0031]Using ...
Embodiment 2
[0032] Example 2: High expression of functionalized amyloid fiber curli (csgA-LACQCL)
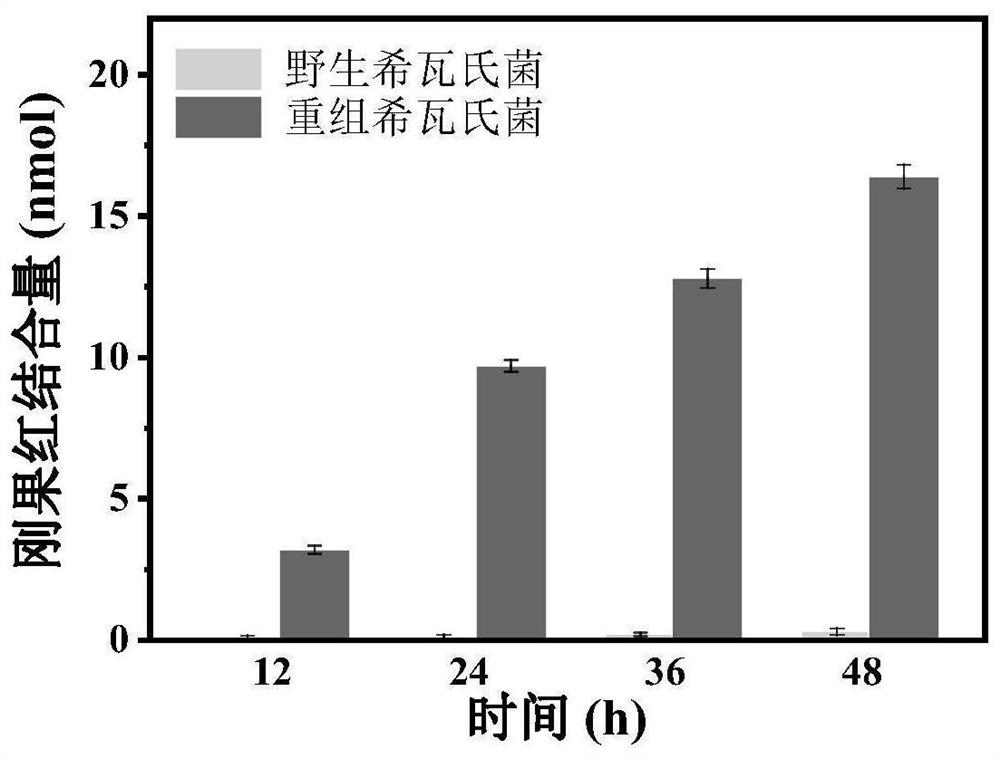
[0033] In this example, the expression of amyloid protein was identified by Congo red (CR) staining, and the concentration of amyloid protein fibers was determined by analyzing the amount of CR binding.
[0034] Centrifuge 1 mL of the wild Shewanella culture and the recombinant Shewanella culture obtained in Example 1 at different incubation times (12h, 24h, 36h, 48h) (5000 rpm, 10 minutes), centrifuge Suspend the precipitate with 0.025 mmol / L CR (dissolved in pH 7.4 phosphate buffer) at 30°C for 10 minutes, then centrifuge (5000 rpm, 10 min), collect the supernatant, and detect the CR binding amount in the supernatant . The absorbance of CR was measured by a UV-Vis spectrophotometer at a wavelength of 490 nm. Compared with the absorbance of 0.025 mmol / L CR at 490 nm wavelength, the CR binding amount of wild Shewanella and recombinant Shewanella was calculated.
[0035] figure 2 It is ...
Embodiment 3
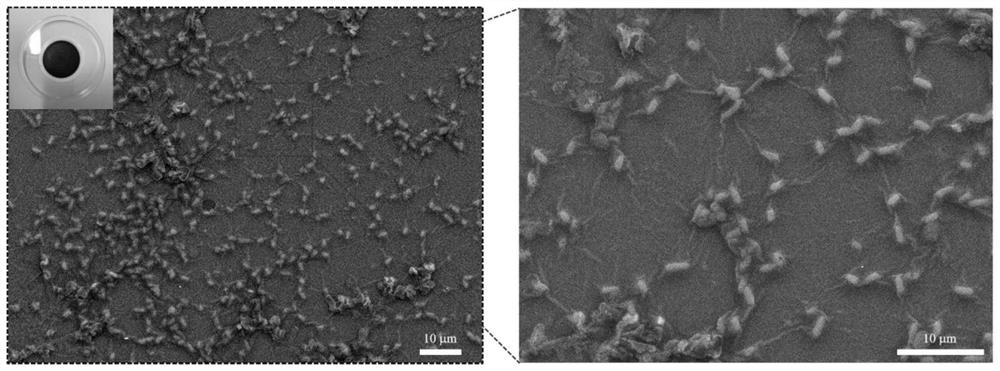
[0036] Embodiment 3: Preparation of recombinant Shewanella adsorption membrane
[0037] In this example, a recombinant Shewanella adsorption membrane of functionalized amyloid fiber curli (csgA-LACQCL) was prepared, and the adsorption membrane included recombinant Shewanella and activated carbon. Concrete preparation method comprises the following steps:
[0038] (1) Inoculate the recombinant Shewanella prepared in Example 1 into YESCA medium at a ratio of 1%, and culture it on a shaker at 30°C at 200rpm to OD 600 =2;
[0039] (2) After cultivation, add 700~800mg of activated carbon powder per 1L of bacterial liquid, soak for 10~12h, and then filter through a water-based microporous membrane with a pore size of 0.45mm to obtain a recombinant expressing functionalized amyloid fiber curli (csgA-LACQCL) Shewanella Absorbent Membrane.
[0040] image 3 is the SEM picture of the prepared recombinant Shewanella adsorption membrane; in the figure, the right picture is a partial e...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com