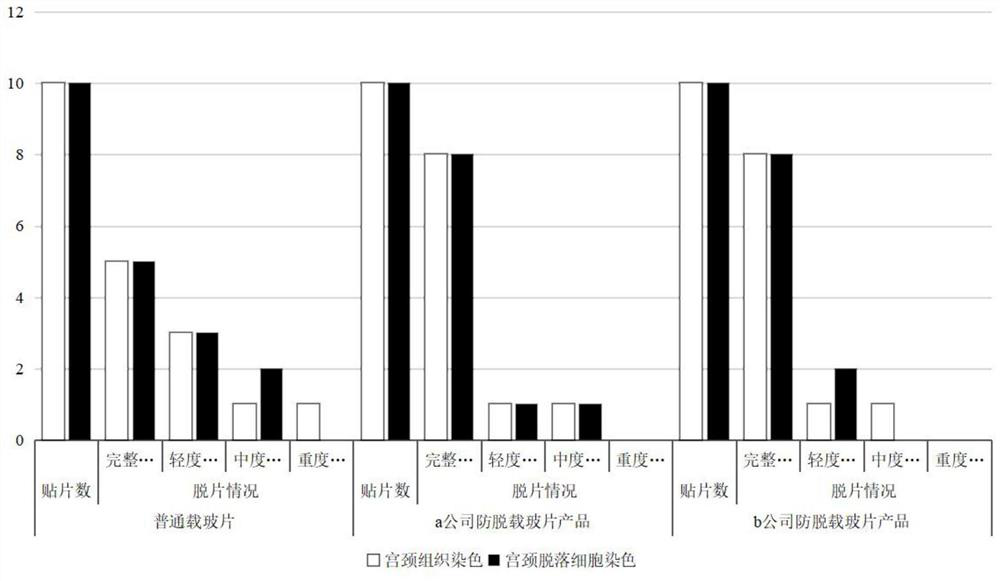
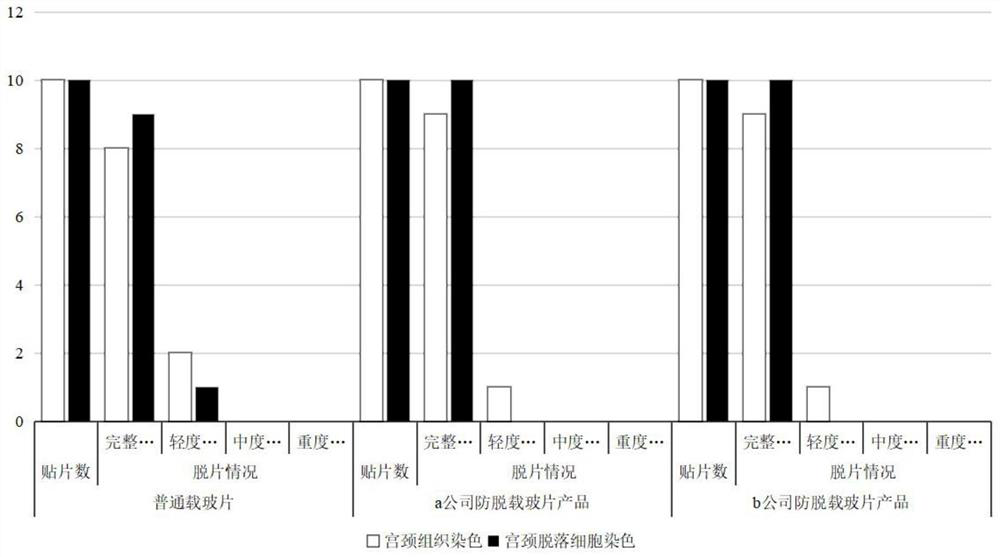
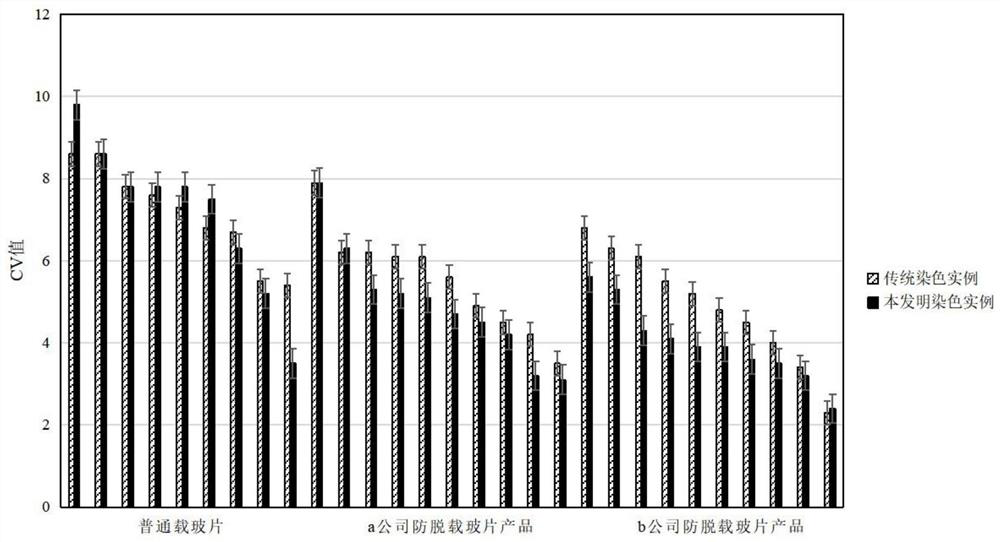
DNA quantitative staining method for anti-slide-off cells
A staining method and cell technology, which is applied in the field of DNA quantitative staining of anti-dropping cells, can solve the problems of unresolved influence of anti-dropping glass slides, reduced accuracy, and cures the symptoms rather than the root causes, and achieves high commercial value and guarantees. Completeness and wide applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] A method for quantitative staining of cell DNA in anti-falling sheets, comprising the following steps:
[0064] I, film production: gradient centrifugal sedimentation method, the steps are as follows:
[0065] (1) The sample is confirmed and numbered accordingly.
[0066] (2) Place the sample bottle on the shaker and shake it fully for 5 minutes.
[0067] (3) Add 2.5 ml of separation liquid (Hunan Pinsheng Biotechnology Co., Ltd., sample gradient separation liquid) into the settling chamber along the wall of the settling chamber or the wall of the filter cup.
[0068] (4) Draw 2.5ml of sample-preservation solution (Hunan Pinsheng Biotechnology Co., Ltd., liquid-based cell preservation solution) and add it along the filter cup wall.
[0069] (5) Put the symmetrical balance into a centrifuge (Hunan Pinsheng Biotechnology Co., Ltd., gradient centrifuge 4000A), centrifuge at 1200 rpm for 3 minutes.
[0070] (6) Remove the filter screen, discard the supernatant, dry it ge...
Embodiment 2
[0090] A method for quantitative staining of cell DNA in anti-falling sheets, comprising the following steps:
[0091] I, film production: gradient centrifugal sedimentation method, the steps are as follows:
[0092] (1) The sample is confirmed and numbered accordingly.
[0093] (2) Place the sample bottle on the shaker and shake it fully for 3 minutes.
[0094] (3) Add 2 ml of separation solution (Hunan Pinsheng Biotechnology Co., Ltd., sample gradient separation solution) into the settling chamber along the wall of the settling chamber or the wall of the filter cup.
[0095] (4) Draw 2ml of sample-preservation solution (Hunan Pisen Biotechnology Co., Ltd., liquid-based cell preservation solution) and add it along the wall of the filter cup.
[0096] (5) Put the symmetrical balance into a centrifuge (Hunan Pinsheng Biotechnology Co., Ltd., gradient centrifuge 4000A), 800 rpm, and centrifuge for 5 minutes.
[0097] (6) Remove the filter screen, discard the supernatant, dry ...
Embodiment 3
[0117] A method for quantitative staining of cell DNA in anti-falling sheets, comprising the following steps:
[0118] I, film production: gradient centrifugal sedimentation method, the steps are as follows:
[0119] (1) The sample is confirmed and numbered accordingly.
[0120] (2) Place the sample bottle on the shaker and shake it fully for 4 minutes.
[0121] (3) Add 1 ml of separation solution (Hunan Pinsheng Biotechnology Co., Ltd., sample gradient separation solution) into the settling chamber along the wall of the settling chamber or the wall of the filter cup.
[0122] (4) Draw 1ml of sample-preservation solution (Hunan Pinsheng Biotechnology Co., Ltd., liquid-based cell preservation solution) and add it along the wall of the filter cup.
[0123] (5) Put the symmetrical balance into a centrifuge (Hunan Pinsheng Biotechnology Co., Ltd., gradient centrifuge 4000A), centrifuge at 2000 rpm for 1 min.
[0124] (6) Remove the filter screen, discard the supernatant, dry it g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com