Recombinant escherichia coli for efficiently producing succinic acid and construction method thereof
A technology for recombining Escherichia coli and Escherichia coli, which is applied in the field of bioengineering, can solve the problems of cofactor metabolic imbalance, metabolic imbalance, product yield and low production intensity, and achieve the effect of increasing production and reducing the accumulation of by-products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
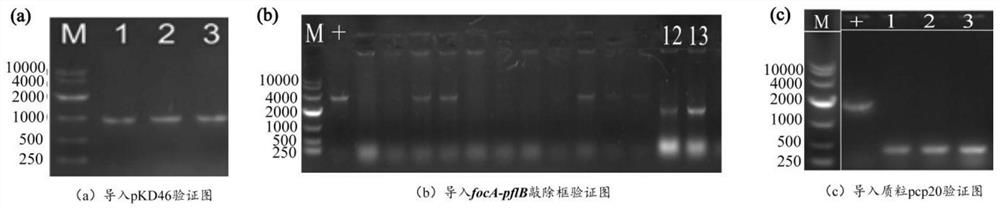
[0046] Example 1: Knockout of the gene encoding pyruvate formate lyase
[0047] (1) In order to knock out the gene encoding pyruvate formate lyase to reduce the amount of acid added by the by-product, a genome editing fragment was constructed with the help of Red homologous recombination technology. The gene editing fragments include upstream and downstream homology arm regions, and resistance screening cassettes. Using the plasmid pKD4 as a template, design sequences such as SEQ ID NO.6 / SEQ ID NO.7 primer pair pflB-focA-S / pflB-focA-A to amplify the resistance screening gene Kan to obtain the pflB-focA knockout frame fragment.
[0048] (2) Transform the pKD46 plasmid into the expression host E.coli FMME-N-5 competent cells, and obtain the recombinant strain E.coli FMME-N-5-pKD46 by colony PCR screening, and then knock out the obtained pflB-focA The frame fragment was transferred into competent cells of the recombinant strain E.coli FMME-N-5-pKD46 by electroporation, and posit...
Embodiment 2
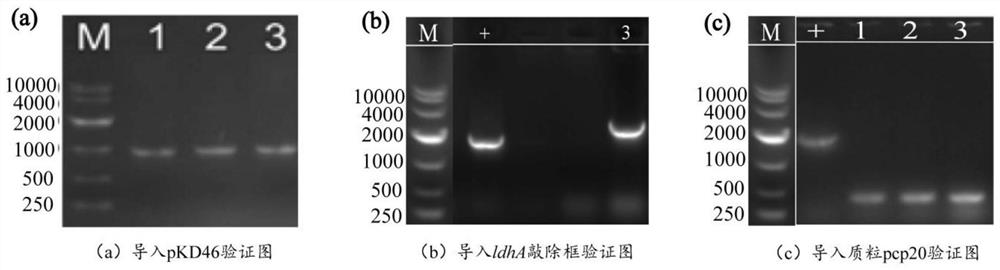
[0052] Example 2: Knockout of lactate dehydrogenase expression gene
[0053] (1) The construction of E.coli FMME-N-5 (ΔfocA-pflB) is the same as in Example 1
[0054] (2) In order to further reduce the amount of by-product lactic acid to knock out the gene encoding lactate dehydrogenase, a genome editing fragment was constructed with the help of Red homologous recombination technology. The gene editing fragments include upstream and downstream homology arm regions, and resistance screening cassettes. Using the plasmid pKD4 as a template, design sequences such as SEQ ID NO.8 / SEQ ID NO.9 primer pair ldhA-S / ldhA-A amplification resistance screening gene Kan to obtain the ldhA knockout frame fragment.
[0055] (3) Transform the pKD46 plasmid into the expression host E.coli FMME-N-5-ΔfocA-pflB competent cells, obtain the recombinant strain E.coli FMME-N-5-ΔfocA-pflB-pKD46 by colony PCR screening, and then The obtained ldhA knockout frame fragment was transferred into competent ce...
Embodiment 3
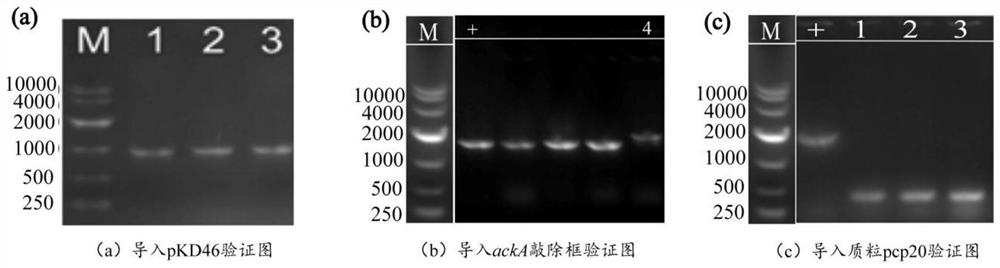
[0059] Example 3: Knockout of Phosphotransacetylase Expression Gene
[0060] (1) The construction of E.coli FMME-N-5 (ΔfocA-pflB-ΔldhA) is the same as in Example 2
[0061] (2) In order to further reduce the amount of by-product acetic acid and ensure cell growth, the gene encoding the phosphoacetyl group was knocked out, and the genome editing fragment was constructed with the help of Red homologous recombination technology. The gene editing fragments include upstream and downstream homology arm regions, and resistance screening cassettes. Using the plasmid pKD4 as a template, design sequences such as SEQ ID NO.10 / SEQ ID NO.11 primer pair pta-S / pta-A amplification resistance screening gene Kan to obtain the pta knockout frame fragment.
[0062] The pKD46 plasmid was transformed into the expression host E.coli FMME-N-5-ΔfocA-pflB-ΔldhA competent cells, and the recombinant strain E.coli FMME-N-5-ΔfocA-pflB-ΔldhA-pKD46 was obtained by colony PCR screening. Then, the obtained p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com