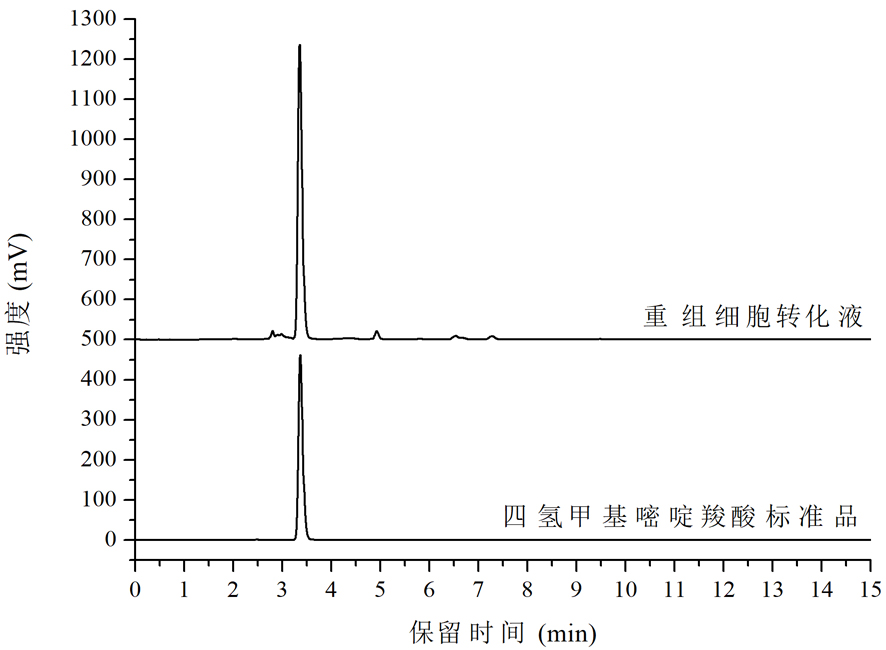
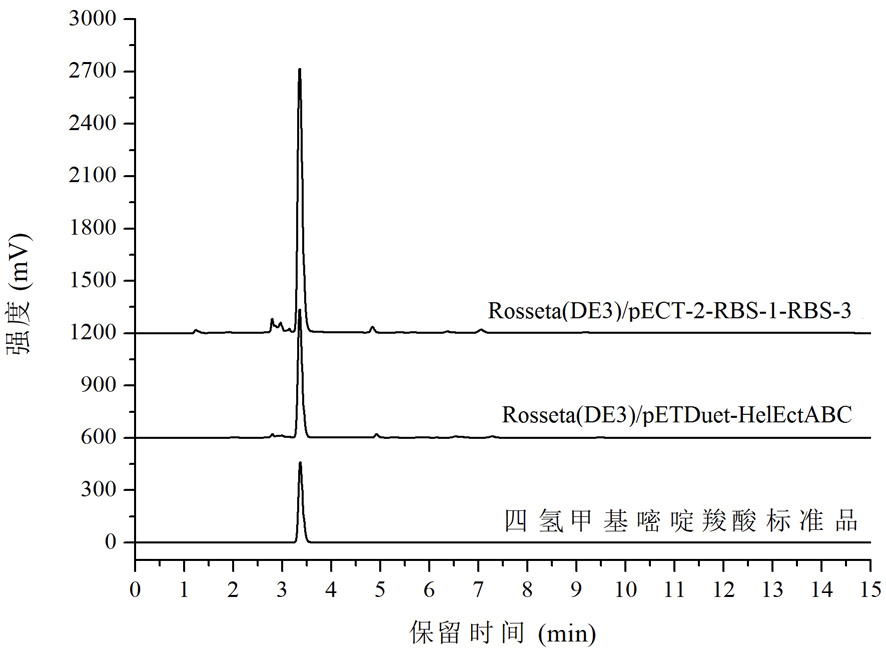
Tetrahydromethylpyrimidine carboxylic acid biosynthetic gene and application thereof
A tetrahydromethylpyrimidine carboxylic acid and biosynthesis technology, applied in the field of genetic engineering and compound biotechnology production, can solve the problems of poor stereospecificity, difficult separation and extraction, and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Codon optimization and total gene synthesis of tetrahydromethylpyrimidinecarboxylic acid biosynthesis gene
[0053] The present invention is based on diaminobutyric acid-2-oxoglutarate aminotransferase (EC 2.6.1.76) and L-2,4-diaminobutyric acid transacetylase (EC 2.3.1.178) derived from Halomonas elongatus and tetrahydromethylpyrimidinecarboxylate synthase (EC 4.2.1.108) (the ID numbers of the three enzymes in the protein database are O52250, O52249 and O52251 respectively), using the degeneracy of codons to encode the above four Design and optimize the codon preference, GC content, and spatial structure of the mRNA of the hydromethylpyrimidine carboxylic acid biosynthetic enzyme, entrust Sangon Bioengineering (Shanghai) Co., Ltd. to carry out the whole gene synthesis, and obtain the nucleotide sequence The polynucleotide encoding diaminobutyrate-2-oxoglutarate aminotransferase as shown in SEQ ID NO: 1, the nucleotide sequence as shown in SEQ ID NO: 2 encodi...
Embodiment 2
[0054] Example 2: Construction of tetrahydromethylpyrimidinecarboxylic acid biosynthetic gene expression vector
[0055] (1) Synthesize two primers respectively having the nucleotide sequences shown in SEQ ID NO: 7 and SEQ ID NO: 8 in the sequence listing. Nucleotide sequence such as SEQ ID NO: 7 and the 5'-end of the primer shown in SEQ ID NO: 8 are set respectively Nde I and xho I Restriction sites and their protective bases. Using the polynucleotide whose nucleotide sequence is shown in SEQ ID NO: 1 as a template, the polymerase chain reaction (PCR) was carried out using the above two primers SEQ ID NO: 7 and SEQ ID NO: 8. The DNA polymerase is Phanta from Nanjing Novizan Biotechnology Co., Ltd. ® Super-Fidelity DNA polymerase. The PCR amplification program was: 95°C for 5min; 94°C for 45s, 56°C for 45s, 72°C for 2min, a total of 30 cycles; 72°C for 10min, then drop to 10°C. PCR products were detected by agarose gel electrophoresis. Under the irradiation of ultrav...
Embodiment 3
[0058] Example 3: Construction of tetrahydromethylpyrimidinecarboxylic acid biosynthetic gene cluster
[0059] 3.1. Tetrahydromethylpyrimidinecarboxylic acid biosynthesis gene cluster P(T7)- lacO - Construction of RBS-2-RBS-1-RBS-3-Ter (T7)
[0060] (1) Synthesize two primers respectively having the nucleotide sequences shown in SEQ ID NO: 13 and SEQ ID NO: 14 in the sequence listing. Nucleotide sequences such as SEQ ID NO: 13 and the 5'-ends of the primers shown in SEQ ID NO: 14 are set respectively Nhe I and Speech I Restriction sites and their protective bases. The expression vector pECT-1 was used as template for PCR amplification. The PCR amplification procedure is the same as (1) in Example 2. The PCR amplified products were detected by agarose gel electrophoresis, separated, and recovered by gel cutting. Nhe I and Speech I double enzyme digestion, using T4 DNA ligase (purchased from Bao Biological Engineering (Dalian) Co., Ltd. (TaKaRa)) to connect the sam...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com