Primer group and kit for simultaneously detecting multiple pathogenic fungi
A primer set and kit technology, applied in the field of microbial detection, can solve the problems of low detection throughput, long detection time, complicated operation, etc., and achieve the effects of high detection sensitivity, fast detection speed, and high detection efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1. Molecular marker screening and primer sequence construction
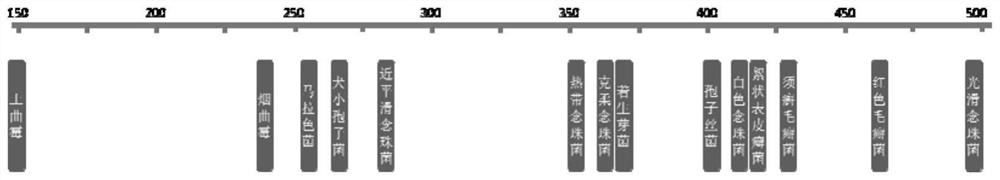
[0043] Collect Candida albicans, Candida tropicalis, Candida glabrata, Candida parapsilosis, Candida krusei, Aspergillus fumigatus, Aspergillus terreus, Trichophyton mentagrophytes, Trichophyton rubrum, Epidermophyton flocculus, Sporothrix, ITS sequences, β-tubulin gene sequences, translation elongation factor (translation elongation factor, TEF), actin (actin) genes, mitochondrial cytochrome B genes, etc. Species molecular marker sequence, and compare the base difference information of the same molecular marker sequence of fungi of different species. Finally, 14 conserved sequence regions with large inter-species variability but intra-species specificity were screened out as targets for species identification. The ITS sequences of Candida, Trichophyton, and Epidermophyton flocculus, Sporothrix, Microsporum canis, Epiphyllum, and Malassezia, and the β-tubulin sequences of Aspergillus Large and sp...
Embodiment 2
[0045] Example 2. Screening of fluorescently labeled primers and establishment of a detection system
[0046] (1) Extract fungal DNA by phenol-chloroform method
[0047] Use an inoculation loop to scrape the fungi from the fungal culture medium, be careful not to scrape the culture medium, grind with liquid nitrogen 4-6 times, and then use the phenol-chloroform method to extract the fungal DNA. Add 500 μL of STE buffer (10 mM, pH 8.0), 50 μL of 10% SDS, 50 μL of proteinase K in sequence, and bathe in water at 56° C. for 6 h. Add 500 μL of phenol-chloroform / isoamyl alcohol (25:24:1), shake and mix, and centrifuge at 12000 rpm for 10 min. Transfer the supernatant to a new centrifuge tube, add 500 μL of chloroform / isoamyl alcohol (24:1), shake and mix, and centrifuge at 12000 rpm for 10 min. Transfer the supernatant to a new centrifuge tube, add 2 times the volume of absolute ethanol, and centrifuge at 12,000 rpm for 10 min. Pour off the supernatant, add 50 μL TE buffer to dis...
Embodiment 3
[0056] Embodiment 3. Sensitivity test
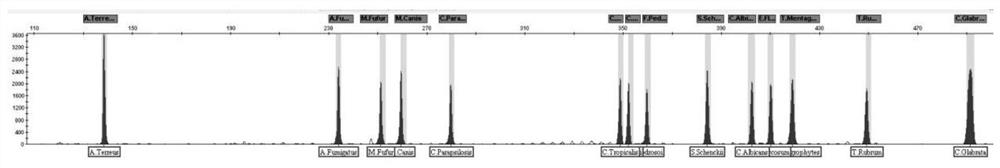
[0057] Dilute the DNA concentration of 14 kinds of pathogenic fungi to 0.1ng / μL, 0.01ng / μL, 0.005ng / μL, 0.002ng / μL, 0.001ng / μL, 0.0005ng / μL, 0.0002ng / μL as provided in Example 2 The multiple detection kit of the pathogenic fungus is detected, and the results are shown in Table 2. The detection sensitivity of the multiple fluorescent-labeled PCR detection system to 14 kinds of fungi is 10 2 copies.
[0058] Table 2 Sensitivity results of multiple detection kits
[0059]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com