High-yield glutathione pichia pastoris strain G3-SA and application thereof
A technology of G3-SA, yeast strain, applied in application, peptide, fermentation and other directions, can solve the problems of unrealistic production scale, can not increase production needs, expensive and other problems, and achieve the effect of enhancing energy supply
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
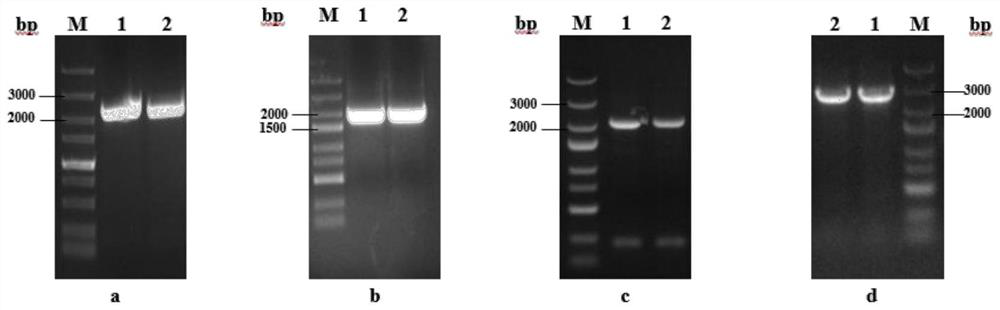
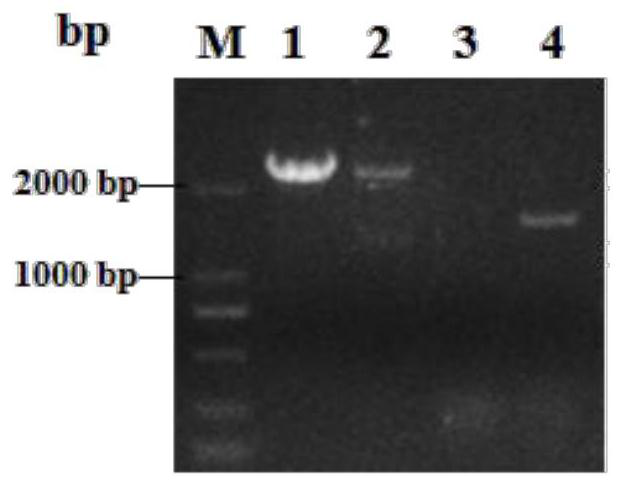
[0038] Embodiment 1: G3 strain construction
[0039] P3 plasmid construction: this patent is constructed with P GAP for the promoter, T AOX1 A constitutive expression strain for the terminator. First, use GAP-F / GAP-R (see Table 1 for the sequence) as primers to amplify the P gene on the genome. GAP promoter sequence. Digest pPIC 3.5K with SacI and BamHI to remove P AOX1 fragment and use P GAP Sequence substitution resulted in a constitutively expressed plasmid, which was named pPICKT. Using the genome of S.cerevisiae BY4741 as a template, GSH1F / GSH1R (see Table 1 for the sequence) and GSH2F / GSH2R (see Table 1 for the sequence) were used to amplify the target gene Scgsh1 and Scgsh2 fragments by PCR respectively (the gene sequence is shown in SEQ.NO.01 , shown in SEQ.NO.02) were respectively connected to pPICKT to form P1 and P2 plasmids. And use YZGF / YZGR (see Table 1 for the sequence) to do the next construction after verification. Then use G1F / G1R (see Table 1 for the...
Embodiment 2
[0041] Embodiment 2: G3-SA strain construction
[0042] Construction of pGAPZT-Scadk1 plasmid: In this study, we used pGAPZA as the starting plasmid, modified it, and named the transformed plasmid pGAPZT. First, use ZXF / ZXR (see Table 1 for the sequence) as primers and pGAPZA as a template to carry out PCR amplification, so that the recovered product X fragment has an XbaI at the front end of the promoter and an NheI restriction site at the end of the terminator. Use BglⅡ and BamHI to digest pGAPZA to remove the middle fragment, and replace it with the X fragment to obtain a plasmid containing the same tail enzyme, which is named p GAP ZX.
[0043] Using ZTF / ZTR as upstream and downstream primers and pPICZA as a template for PCR amplification to obtain P AOX1 Fragment, use BglⅡ and XbaΙ to digest the pGAPZX vector and combine with P AOX1 Fragments ligated to obtain a AOX1 The plasmid, named pGAPZT, can integrate the expression cassette of single or multiple target genes in...
Embodiment 3
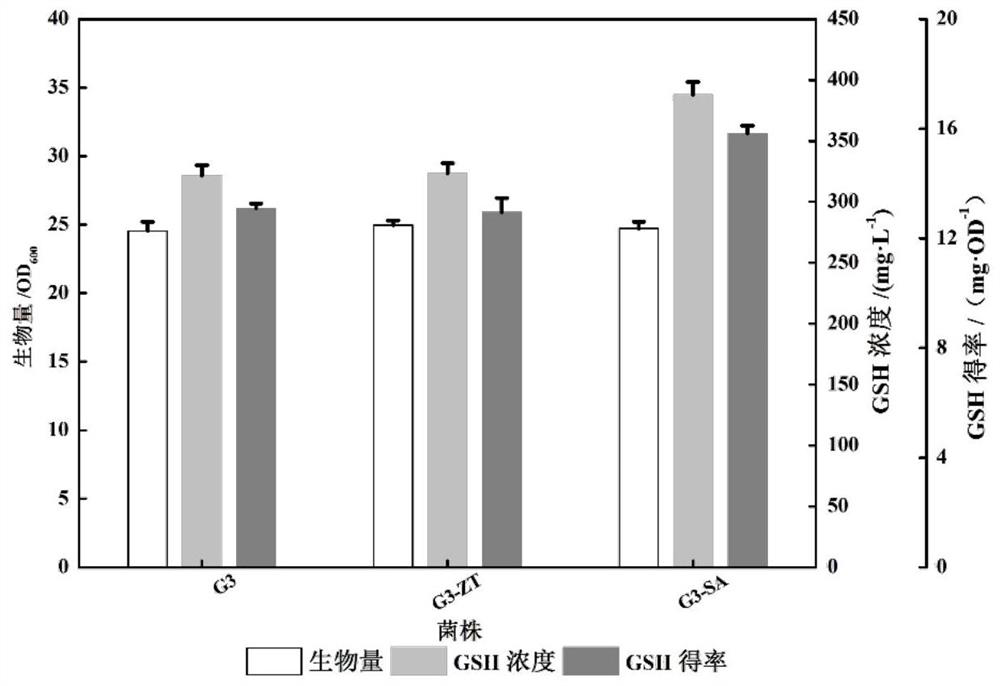
[0048] Embodiment 3: G3-SA bacterial strain shakes flask fermentation
[0049] The fermentation medium at shake flask level is YPD medium, including: glucose 20g / L, yeast powder 10g / L, peptone 20g / L.
[0050] Shake flask fermentation: the seed culture medium is YPD medium and the fermentation medium is YPD medium. Pick the strain from the glycerol tube to the YPD plate, culture at 30°C for 2-3 days, pick the grown single colony into a shake flask containing 10mL seed medium, culture at 30°C for 16-18h, and then set the initial OD as 0.2 Inoculated into the fermentation medium and cultured at 220 rpm for 30 h, and initially added a mixed solution with concentrations of 10 mM glutamic acid, 10 mM cysteine, and 10 mM glycine. After 30 hours of fermentation, the bacteria were collected, and the supernatant was used to measure ethanol, glycerol and glucose. Incubate with 40% ethanol solution at 220rpm for 2h. After centrifugation, the supernatant contains high concentration of GS...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com