Kit and method for rapidly detecting archaea
A kit and rapid technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of low frequency of use, high sequencing error rate, long detection cycle, etc., to achieve specificity Strong, low experimental cost, easy to operate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] 1. A primer set and DNA fragments for rapid detection of archaea by chemical synthesis.
[0037] Among them, the member sequences of the primer set are the sequences shown in SEQ ID NO: 1, SEQ ID NO: 2, and SEQ ID NO: 3; the sequences of all DNA fragments are SEQ ID NO: 4, SEQ ID NO: 5, and SEQ ID NO: 6. The sequences shown in SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, and SEQ ID NO: 10.
[0038] 2. The chemically synthesized DNA fragments were amplified by PCR using the primer pair SEQ ID NO: 1 (upstream primer) / SEQ ID NO: 3 (downstream primer), and the amplified product was used as a standard for the sample to be tested.
[0039] The PCR reaction system is: 10*EasyTaq Buffer: 5 μL, dNTPs (2,5 mM): 4 μL, upstream primer (10 μmol / L): 1 μL, downstream primer (10 μmol / L): 1 μL, EasyTaq DNA Polymerase: 1 μL, template DNA ( Chemically synthesized DNA fragments): 1 μL, ddH20: 37 μL;
[0040] The PCR reaction program was: 94°C for 5min; 30 cycles of 94°C for 30s, 45-55°C for...
Embodiment 2
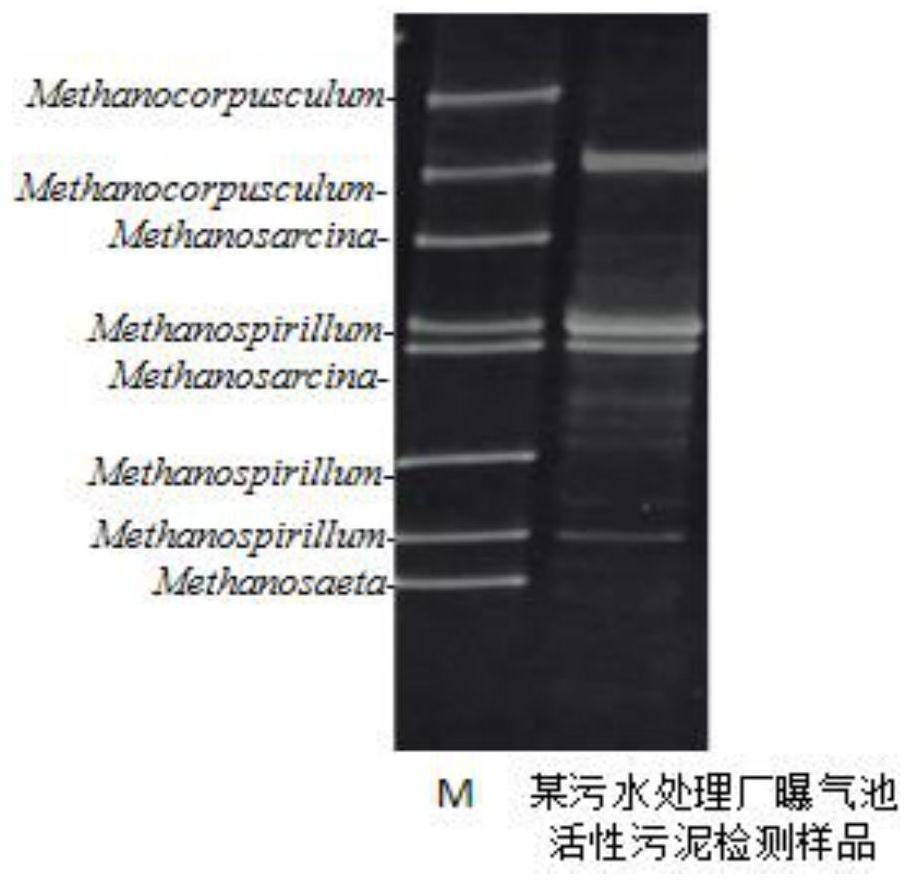
[0044] The method for detecting the activated sludge archaea in the aeration tank of a sewage treatment plant: comprises the following steps:
[0045] 1. Extract the total DNA of the microbial genome from the activated sludge in the aeration tank
[0046] (1) Dilute and dissolve the activated sludge with ultrapure water, filter it with a 0.22 μm filter membrane, put the filter membrane with microorganisms into an EP tube, wash it twice with STE buffer, and then centrifuge at 12000 rpm for 1 minute.
[0047] (2) Add 300 μl of STE buffer solution and 30 μl of lysozyme, wash and mix with a pipette, and place in a constant temperature water bath at 37° C. for 3.5 hours.
[0048] (3) Add 35 μl of SDS and 5 μl of Proteinase K, and keep in a constant temperature water bath at 55° C. for 2.5 hours.
[0049] (4) Take it out after completion, cool to room temperature, and add 70 μl of NaCl.
[0050] (5) Add 450 μl DNA extraction phenol reagent, mix for 10 minutes, centrifuge at 12000 r...
Embodiment 3
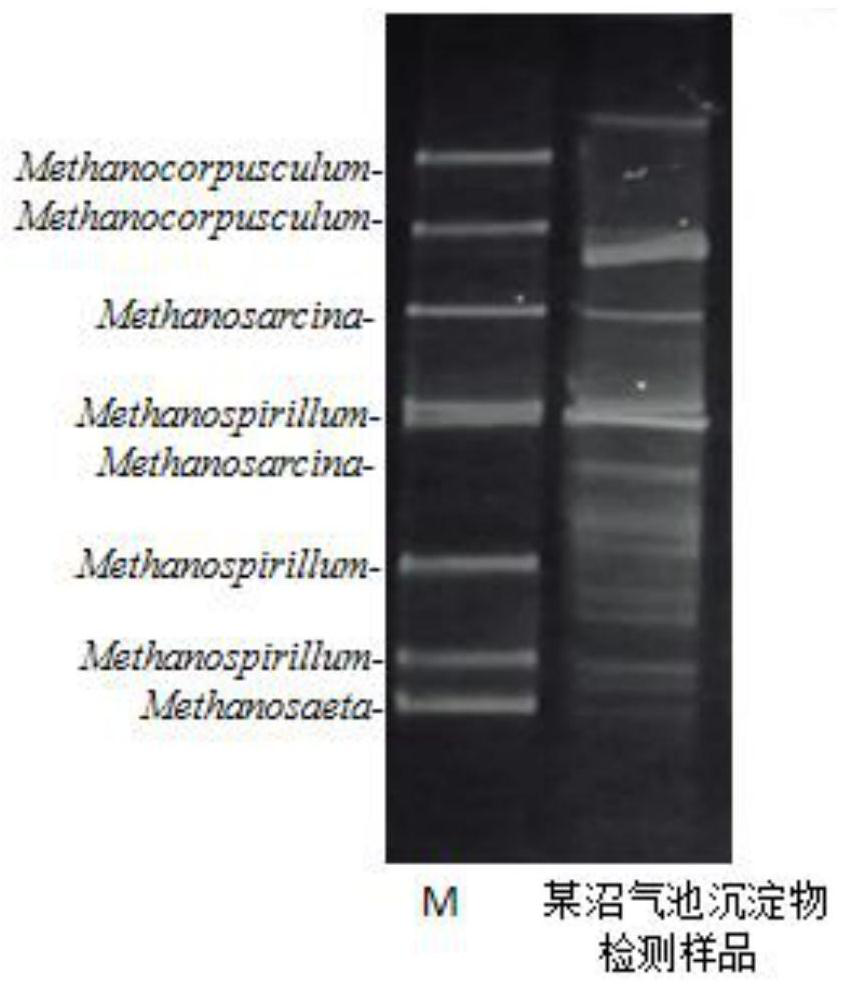
[0066] A method for detecting archaea in sediments of rural biogas digesters: comprising the following steps:
[0067] 1. Extraction of microbial genome total DNA in biogas digester sediment
[0068] 1.1 DNA extraction
[0069] (1) Dilute the solid-liquid sample in the biogas digester with ultrapure water, and then filter it with a 0.22 μm filter membrane, put the filter membrane with archaeal microorganisms into the EP tube, wash it twice with STE buffer, and then Centrifuge at 12000 rpm for 1 min.
[0070] (2) Add 300 μl of STE buffer solution and 30 μl of lysozyme, wash and mix with a pipette, and place in a constant temperature water bath at 37° C. for 3.5 hours.
[0071] (3) Add 35 μl of SDS and 5 μl of Proteinase K, and keep in a constant temperature water bath at 55° C. for 2.5 hours.
[0072] (4) Take it out after completion, cool to room temperature, and add 70 μl of NaCl.
[0073] (5) Add 450 μl DNA extraction phenol reagent, mix for 10 minutes, centrifuge at 120...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com