Aldolase mutant and its application in production of 1, 3-propylene glycol
A mutant, phosphate aldolase technology for applications, microbial-based methods, enzymes, etc., capable of solving problems such as restricted 3-HPA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
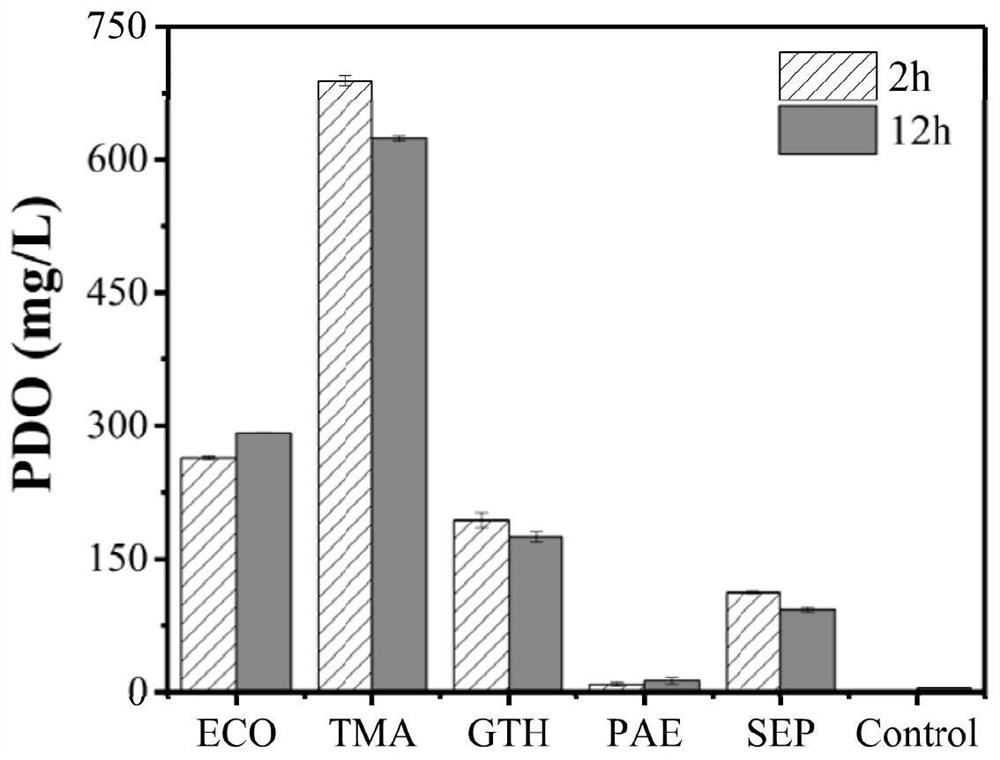
[0054] Example 1: Screening efficient DERA for novel formaldehyde and acetaldehyde to PDO production pathway
[0055]Different sources of DERA were constructed on the expression vector pRSFduet-1-DhaT with the gene encoding DhaT, respectively: deoxyribose-5-phosphate aldolase pRSFduet-DERA derived from Escherichia coli ECO -DhaT; deoxyribose-5-phosphate aldolase pRSFduet-DERA derived from Thermotoga maritima TMA -DhaT(DERA TMA The amino acid sequence is shown in SEQ ID NO.1, and the nucleotide sequence is shown in SEQ ID NO.3); deoxyribose-5-phosphate aldolase pRSFduet-DERA derived from Geobacillus thermodenitrificans GTH -DhaT; Deoxyribose-5-phosphate aldolase pRSFduet-DERA derived from Pyrobaculum aerophilum PAE -DhaT; deoxyribose-5-phosphate aldolase pRSFduet-DERA derived from Staphylococcus epidermidis SEP -DhaT. The above expression vectors were respectively transformed into the E. coli expression host E.coli BL21(DE3) (Control did not express any DERA, only expressed...
Embodiment 2
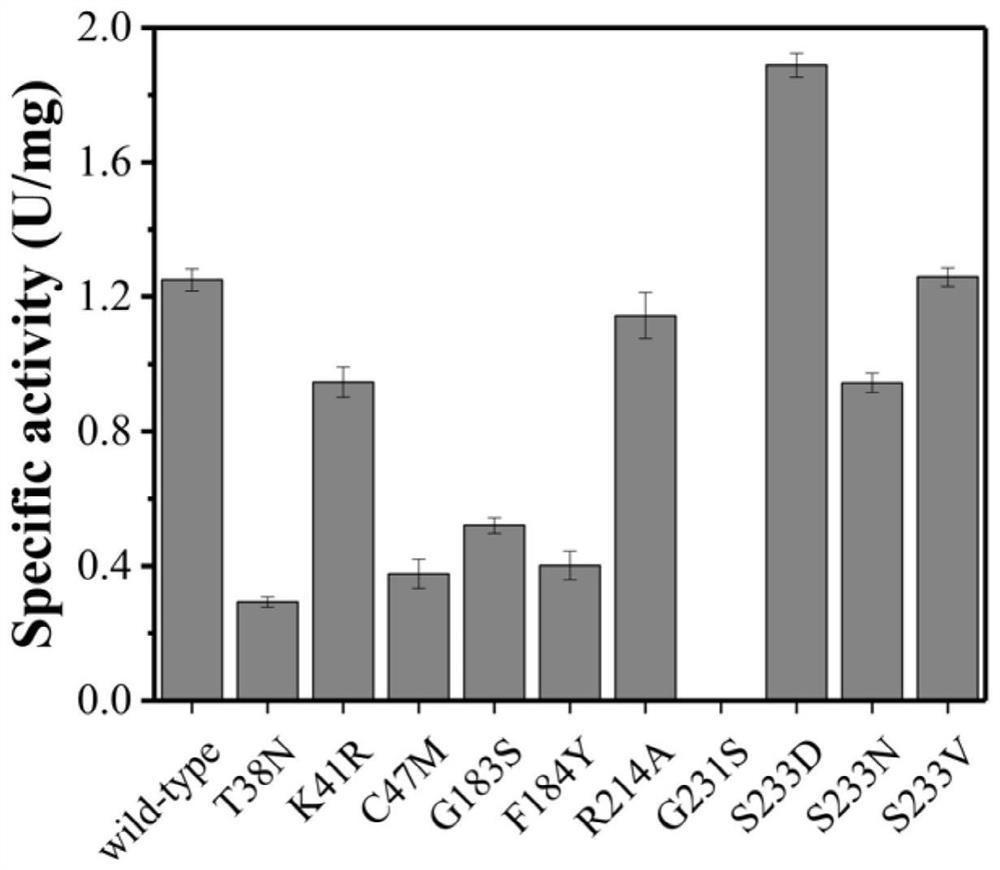
[0057] Example 2: For DERA TMA Enzyme activity assay results for semi-rational design mutations
[0058] From Example 1, we can screen the deoxyribose-5-phosphate aldolase derived from Thermotoga maritima with the best performance in this biotransformation system. Then, we carried out a semi-rational mutation design on the enzyme, and carried out enzyme activity determination on the obtained mutants, the results are as follows: image 3 , its S233D mutant obtained a catalytic activity 51% higher than that of the wild type, and its specific activity was 1.89±0.04U / mg. The amino acid sequence of the S233D mutant is shown in SEQ ID NO.2, and the nucleotide sequence is shown in SEQ ID Shown in NO.4.
Embodiment 3
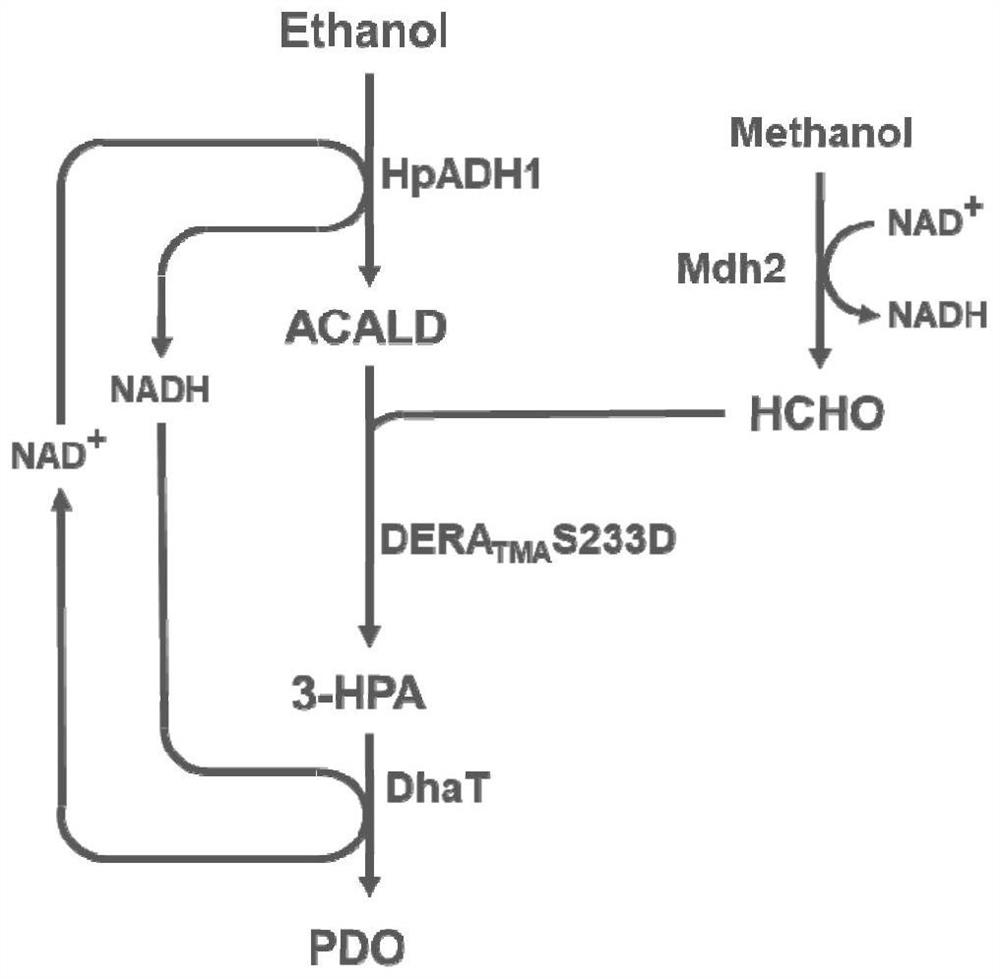
[0059] Example 3: Validation of novel formaldehyde and ethanol production pathways to PDO
[0060] The HpADH1 gene (shown in SEQ ID NO.6) derived from Hansenula polymorpha and the deoxyribose-5-phosphate aldolase mutant DERA derived from Thermotoga maritima TMA The S233D gene (shown in SEQ ID NO.4) and the DhaT gene (shown in SEQ ID NO.5) derived from Klebsiella pneumoniae were constructed on the expression vector pRSFduet-1. The above expression vectors were respectively transformed into the E. coli expression host E.coli BL21(DE3), single clones were picked and cultured in LB medium at 37°C for 8h, and then transferred to 400mL containing 50μg / mL kanamycin LB medium to OD 600 After reaching 0.6-0.8, add 0.2mM IPTG to induce gene expression, continue culturing at 30°C for 10 hours, measure the absorbance value of the bacteria at 600nm, collect a certain amount of bacteria and centrifuge to remove the supernatant, then buffer with a certain amount of 50mM phosphate Resuspend...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com