Nucleic acid sequencing method of nucleoside non-labeled FRET
A technology of nucleic acid sequencing and nucleoside non-labeling, which is applied in the field of nucleic acid sequencing of nucleoside non-labeled FRET, can solve the problem of high error rate of base synthesis, achieve the effect of strong versatility and improve accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
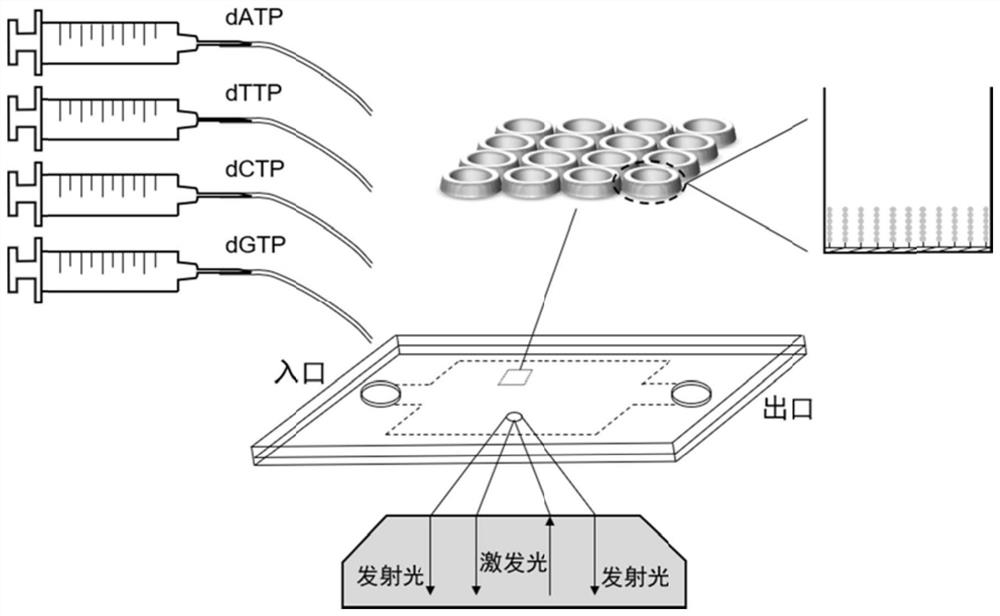
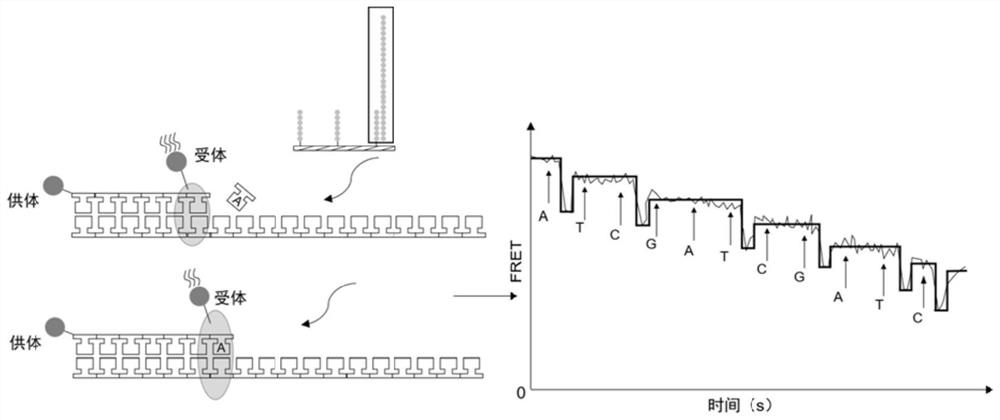
[0032] Preparation and application of microwell arrays: PDMS (polydimethylsiloxane) chips containing countless microwells (2μm×2μm×2μm) were prepared by soft lithography, with more than 500,000 microwells on each chip. And the known sequence is chemically modified on the surface of the chip as a linker to capture the nucleic acid molecule to be tested (6-8 bases of the known sequence are complementary to the linker of the subsequent library construction), and the known sequence is also used as a primer for the synthesis of the complementary strand. The donor (Alexa Fluor 647) and acceptor (Alexa Fluor 555) fluorophores can either be modified at the 5' end of the known sequence, the other can be modified on the DNA polymerase, or both can be modified on the DNA polymerase. Design the nucleic acid sequence according to the known sequence on the chip surface and the A tail added by PCR operation, and complete the connection on the unknown nucleic acid molecule to be tested by DNA ...
Embodiment 2
[0039] On the basis of the sequencing platform in Example 1, a PDMS (polydimethylsiloxane) chip containing micropores of 2 μm×2 μm×2 μm was prepared by soft lithography, with more than 500,000 microwells on each chip, and The known sequence (TCGCCTAT) is chemically modified on the surface of the chip as an adapter to capture the nucleic acid molecule to be tested. Each microwell is modified with countless known sequences, and AlexaFluor 647 is modified at the 5' end of the known sequence. Use the genomic DNA of GM12878 to amplify the HLA-A gene fragment (172bp, NCBI Homo sapiens Gene ID: 3105), 50μL system: Premix Ex Taq HS 25μL, DNA 50ng, forward and reverse primers 0.4μM each (forward primer: 5′ -GGATTACATCGCCCTGAAC-3'; reverse primer: 5'-CGTCTCCTTCCCGTTCTC-3-3'), less than 50 μL was filled with enzyme-free water. The amplification program was: 95°C for 5min; (95°C for 30s, 60°C for 20s, 72°C for 30s) 30 cycles; 72°C for 5min; 4°C for storage. The PCR product is designed wi...
Embodiment 3
[0045]On the basis of the sequencing platform in Example 1, a PDMS (polydimethylsiloxane) chip containing numerous micropores was prepared by soft lithography, each micropore has a diameter of 1 μm and a depth of 2 μm. Contains millions of microwells, and chemically modify known sequences (TCGCCTAT) on the surface of the chip as adapters to capture nucleic acid molecules to be tested. Each microwell is modified with countless known sequences, and the known sequences are also synthesized as complementary strands primers. Use the genomic DNA of GM12878 to amplify the HLA-A gene fragment (172bp), 50μL system: Premix Ex Taq HS 25μL, DNA 50ng, forward and reverse primers 0.4μM each (forward primer: 5′-GGATTACATCGCCCTGAAC-3′; reverse Primer: 5′-CGTCTCCTTCCCGTTCTC-3-3′), less than 50 μL was filled with enzyme-free water. The amplification program was: 95°C for 5min; (95°C for 30s, 60°C for 20s, 72°C for 30s) 30 cycles; 72°C for 5min; 4°C for storage. The nucleic acid sequence (5'-T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com