Saccharomyces cerevisiae engineering bacteria and application thereof in preparation of protocatechuic acid
A strain of Saccharomyces cerevisiae, the technology of Saccharomyces cerevisiae, applied in the field of genetic engineering, can solve the problems of ineffective use of benzene ring structure and waste of resources, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
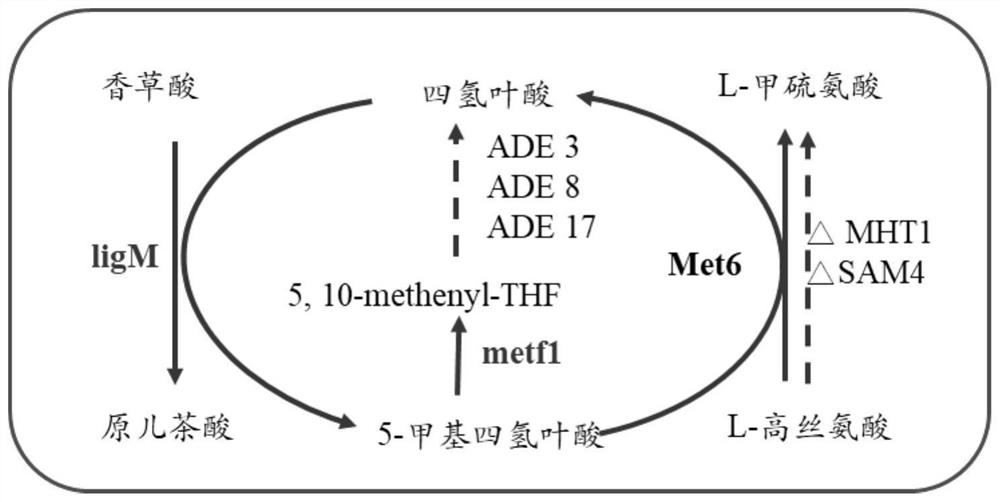
[0074] 1. Construction of strains ZR01-ZR03: Obtain 400 bp sequences of Saccharomyces cerevisiae ADH6 by PCR, name them ADH6L and ADH6R, carry out overlap, and obtain ADH6LR as donor DNA. The gRNA plasmid was constructed according to the ADH6 sequence, the gRNA, cas9, and donor DNA were introduced into Saccharomyces cerevisiae BY4742, and the SC screening plate was spread. The obtained transformants were streaked and purified, and then the yeast genome was extracted for PCR verification and sequencing. According to the above method, ADH7 and BDH2 were sequentially knocked out, named ZR01, and MHT1 was further knocked out, named ZR02. On the basis of ZR02, SAM4 was knocked out and named ZR03.
[0075] 2. Construction of yeast engineering bacteria PCA1, PCA2, PCA3:
[0076] PCA1 (transfer of exogenous gene 4CL, ppech in ZR01): amplification of yeast promoter P by PCR TPI1P terminator T GPM1 , overlap with the gene 4CL gene to obtain the module P TPI1P -4CL-T GPM1 . Amplifi...
Embodiment 2
[0085] The ability of different strains of embodiment 2 to synthesize protocatechuic acid
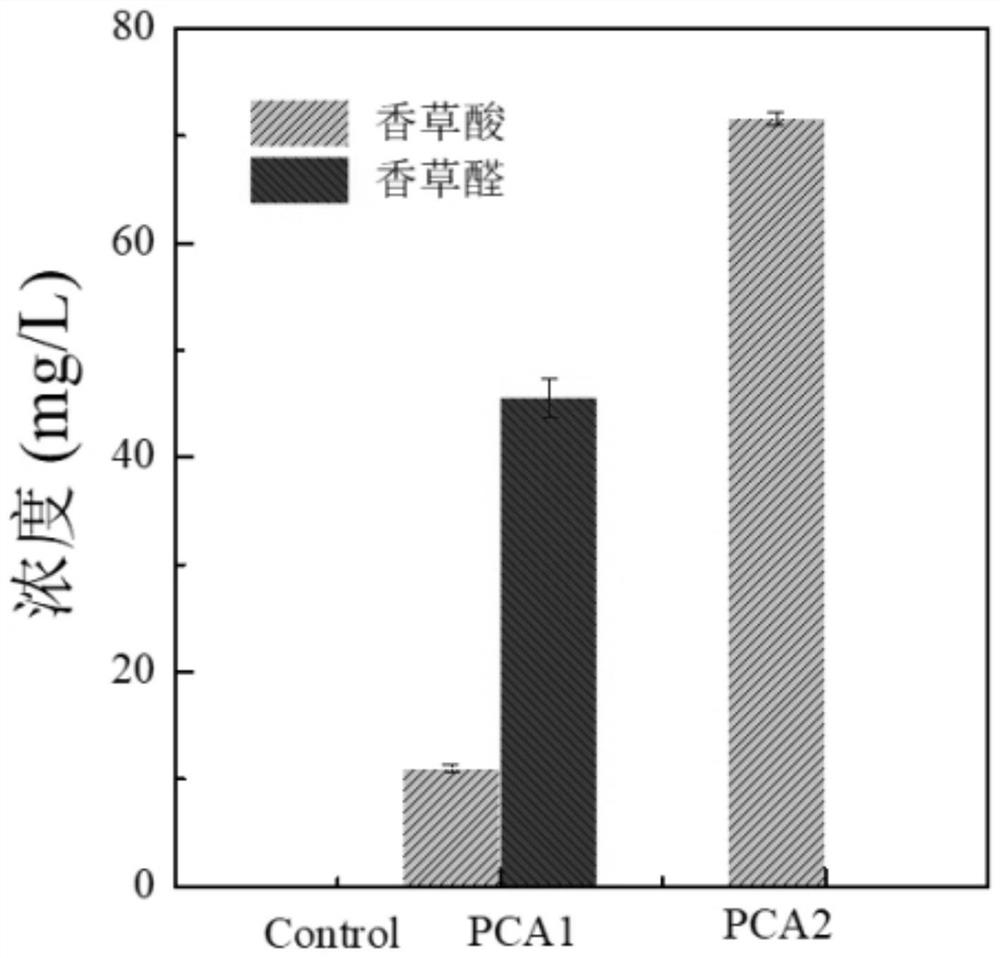
[0086] The strain PCA1 PCA2 constructed in Example 1 was initially added with SC-LEU of 100mg / L ferulic acid (synthetic yeast nitrogen source YNB 6.7g / L, glucose 20g / L, mixed amino acid powder 2g / L lacking leucine) Cultivate under medium for 24h, analyze the output of its vanillic acid and vanillin, the results are shown in figure 1 .
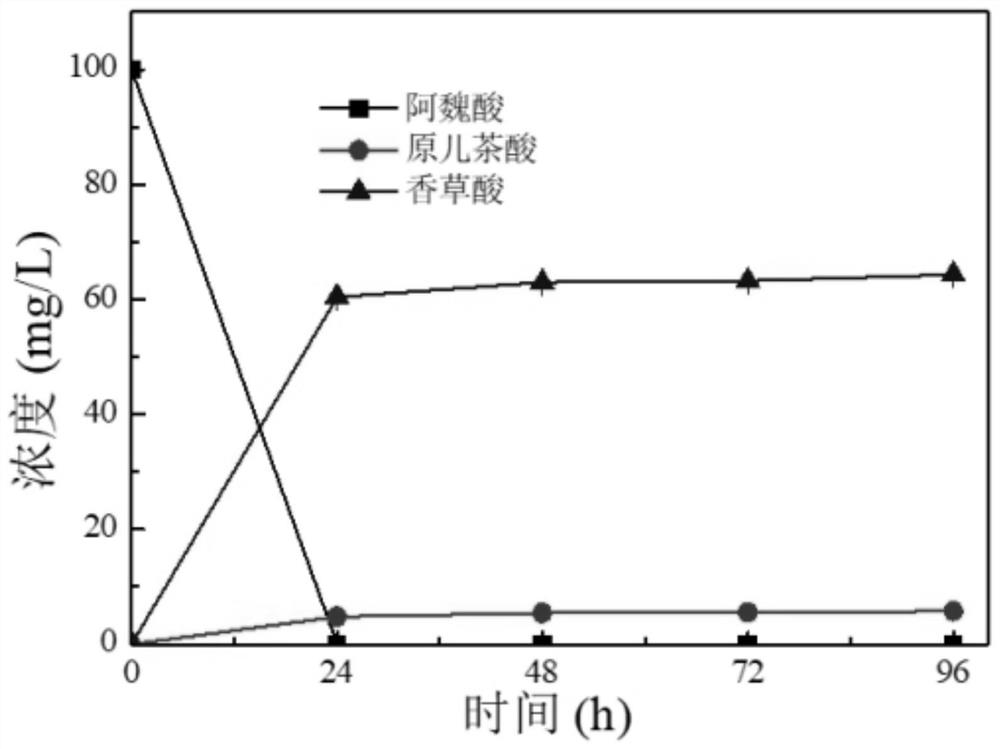
[0087] The strain PCA3 constructed in Example 1 was initially cultured in SC-LEU (synthetic yeast nitrogen source YNB 6.7g / L, glucose 20g / L, leucine-deficient mixed amino acid powder 2g / L) initially added with 100mg / L ferulic acid Production of protocatechuic acid and vanillic acid ( figure 2 ).
[0088] The bacterial strain PCA4, PCA5, PCA6, PCA7, PCA8, PCA9 constructed in Example 1 were cultivated for 24h under the SC medium of the corresponding deficiency of 100mg / L initially added 100mg / L vanillic acid and the production of protocatechuic acid an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com