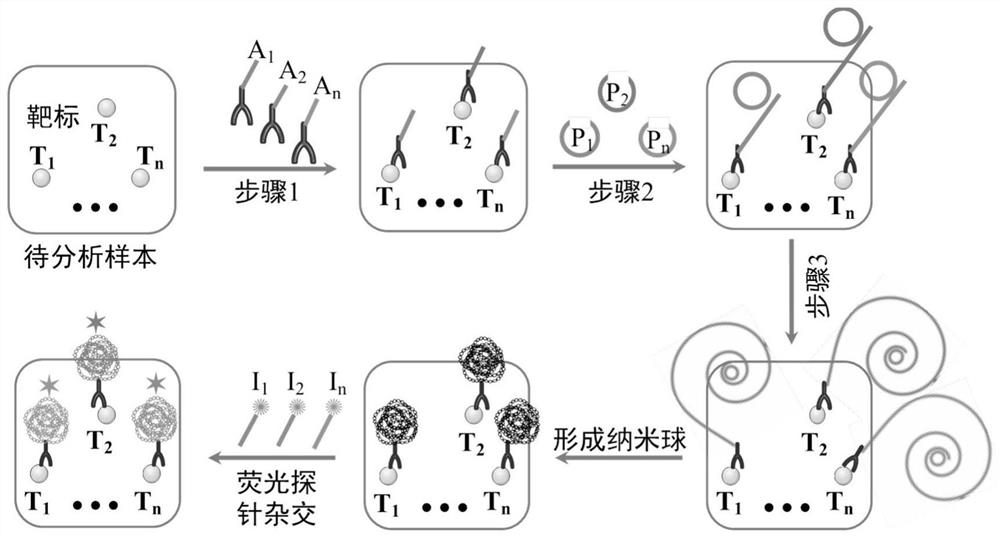
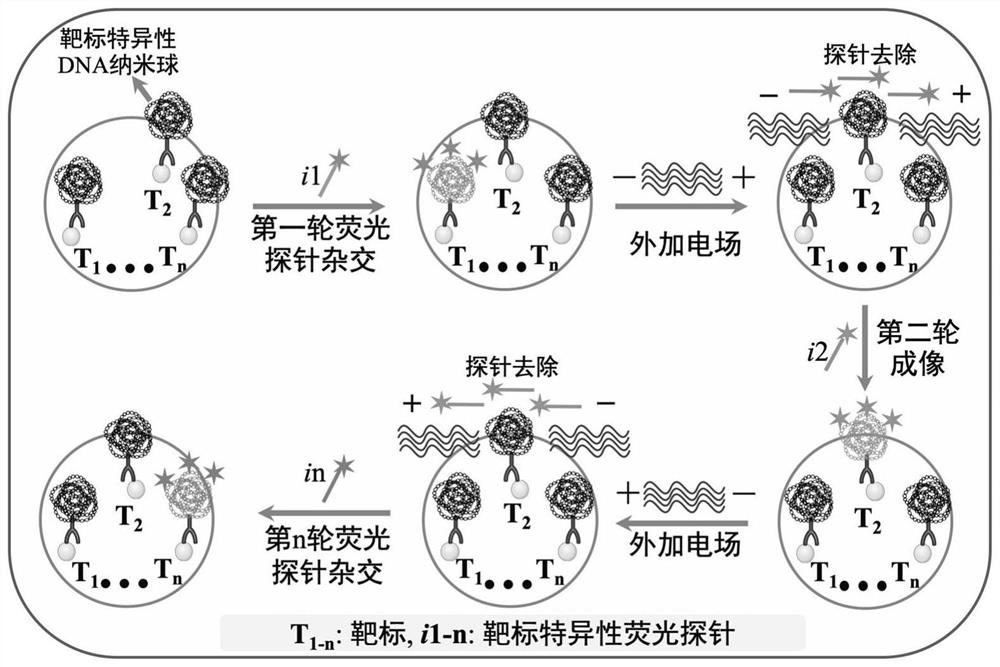
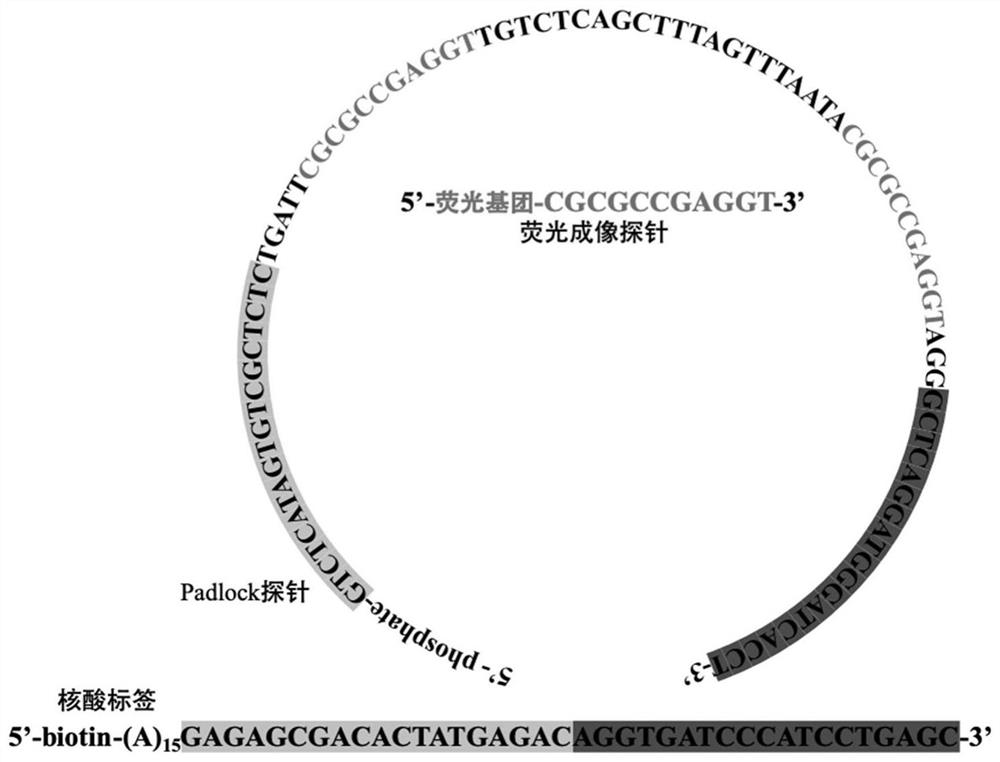
Target positioning and quantitative detection method based on DNA spherical nanostructure imaging
A quantitative detection method and nanostructure technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, and labels used in chemical analysis, etc., can solve the problem of limited, ineffective detection of low-abundance and rare targets, and spontaneous detection of samples. Issues such as fluorescence and light scattering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] (1) Experimental materials and reagents:
[0058] MDA-MB-231 cell line was purchased from Shanghai ATCC cell bank; cell culture grade phosphate buffered saline solution (PBS solution, without calcium chloride, magnesium chloride, 1×PBS pH 7.4) was purchased from the United States DMEM medium (containing penicillin-streptomycin double antibody) was purchased from Keygen; trypsin (Trypsin) was purchased from the United States Sterile fetal bovine serum (FBS) was purchased from Natocor-Industria, Argentina Bovine serum albumin (bovine serum albumin, BSA) was purchased from American Amresco Company; Glass Bottom Cell Culture Dish cell culture dish was purchased from Wuxi Nice Biotechnology Co., Ltd.; biotin, formamide, dimethyl sulfoxide (DMSO) And polyethylene glycol tert-octyl phenyl ether (Triton X-100) was purchased from Sigma-Aldrich Company of the United States; 4′, 6-diamidino-2-phenylindole (DAPI) was purchased from Wuhan Boster (Boster ) company; all oligonucl...
Embodiment 2
[0070] Example 2 Oligonucleotide labeling and corresponding design of single-stranded DNA molecules used to form circular templates
[0071] see image 3 , in this example, see Example 1 for specific experimental materials and reagents, the 5' end of the oligonucleotide tag 1 is biotinylated, so that the tag and the target-specific binding molecule are passed through biotin- Streptavidin system for bridging. The single-stranded DNA molecule (Padlock molecule 1) contains a complementary part to the nucleic acid tag to form a circular template, and also contains a template sequence for the generation of the hybridization sequence of the fluorescently labeled probe 1, so that only when the oligonucleotide tag 1 is amplified In order to generate a complementary hybridization sequence with the fluorescent label probe 1, the specificity of the fluorescent signal is ensured.
Embodiment 3
[0072] Example 3 Glyceraldehyde-3-phosphate dehydrogenase (glyceraldehyde-3-phosphate dehydrogenase) protein detection
[0073] see Figure 4 , in this embodiment, specific experimental materials and reagents, cell culture experimental steps, contents and conditions, oligonucleotide tag-labeled antibody modification experimental steps, contents and conditions refer to Example 1, will be fixed with 4% paraformaldehyde The final MDA-MB-231 cell sample is in contact with the target-specific binding coupler, so that it can be stably and specifically bound to the sample, and the target is formed under the action of rolling circle replication mediated by the nucleic acid label on the target-specific binding coupler. Specific DNA nanospheres can be hybridized with specific fluorescently labeled probes to form hybrids, forming fluorescent bright spots that are clearly distinguished from the background under a fluorescent microscope.
[0074]Target detection experimental steps, conten...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com