Genetically engineered bacterium for efficiently producing (2S, 3S)-2, 3-butanediol, construction method and application thereof
A technology of genetically engineered bacteria and butanediol, applied in genetic engineering, microorganism-based methods, biochemical equipment and methods, etc., can solve control difficulties, high implementation costs, low optical purity of 2,3-butanediol, etc. problem, to achieve the effect of increasing product concentration, reducing accumulation, and improving substrate conversion rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1: Construction of recombinant plasmid pET28a-KbudC / pETDuet-KbudC:
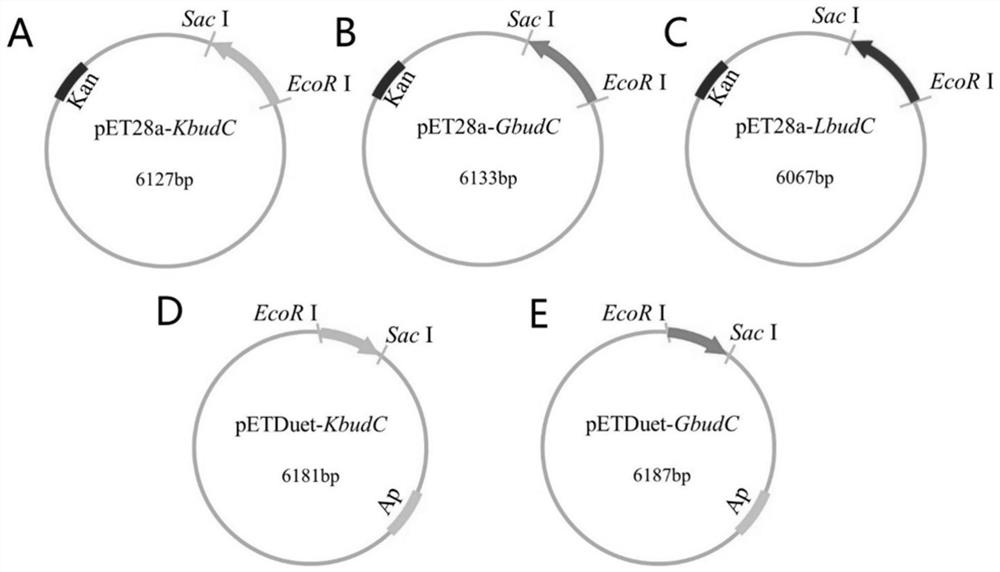
[0059] The 2,3-butanediol dehydrogenase gene KbudC of Klebsiella pneumoniae was cloned into the pET28a plasmid and the pETDuet plasmid to construct the pET28a-KbudC plasmid and pETDuet-KbudC, and the map of the pET28a-KbudC plasmid was as follows figure 1 As shown in A, the map of pETDuet-KbudC plasmid is as follows figure 1 D shows. The specific operation steps are as follows:
[0060]Klebsiella pneumoniae genome was used as a template for PCR amplification, and bioinformatics software was used to design the amplification primers of the gene KbudC sequence, and the gene KbudC sequence was amplified, and the primer sequences were as follows:
[0061] Primer1: 5′-ATGGGTCGCGGATCCGAATTCATGAAAAAAGTCGCACTTGTTACC-3′ (contains EcoRI restriction site),
[0062] Primer2: 5′-GCAAGCTTGTCGACGGAGCTCTTAGTTAAATACCATCCCGCCG-3′ (containing SacI restriction site);
[0063] The PCR amplification system was pr...
Embodiment 2
[0066] Example 2: Construction of recombinant plasmid pET28a-GbudC / pETDuet-GbudC:
[0067] The 2,3-butanediol dehydrogenase gene GbudC of Corynebacterium glutamicum was cloned into the pET28a plasmid and the pETDuet plasmid, and the pET28a-GbudC plasmid and pETDuet-GbudC were constructed. The map of the pET28a-GbudC plasmid is as follows figure 1 As shown in B, the map of pETDuet-GbudC plasmid is as follows figure 1 Shown in E. The specific operation steps are as follows:
[0068] The genome of Corynebacterium glutamicum was used as a template for PCR amplification, and the amplification primers of the gene GbudC sequence were designed by using bioinformatics software, and the gene GbudC sequence was amplified, and the primer sequences were as follows:
[0069] Primer3: 5′-GCAAGCTTGTCGACGGAGCTCATGAGCAAAGTTGCAATGGTTACC-3′ (contains EcoRI restriction site),
[0070] Primer4: 5'-GCAAGCTTGTCGACGGAGCTCTTAGTTGTAGAGCATGCCGCC-3' (containing SacI restriction site);
[0071] The PCR...
Embodiment 3
[0074] Embodiment 3: Construction of plasmid pET28a-LbudC
[0075] The 2,3-butanediol dehydrogenase gene sequence of Staphylococcus aureus was searched at NCBI, which was synthesized by Shanghai Sangon with codon optimization and then ligated into pET28a to obtain the pET28a-LbudC plasmid. The map of the pET28a-LbudC plasmid is as follows figure 1 C shown. Use the pET28a-LbudC plasmid as a template for PCR verification, and the primer sequences are as follows
[0076] Primer5: 5′-ATGGGTCGCGGATCCGAATTCATGAACCTGAAAGATGCGAAAAT-3′
[0077] Primer6: 5′-GCAAGCTTGTCGACGGAGCTCTTACTGCGCCGCCCACGG-3′
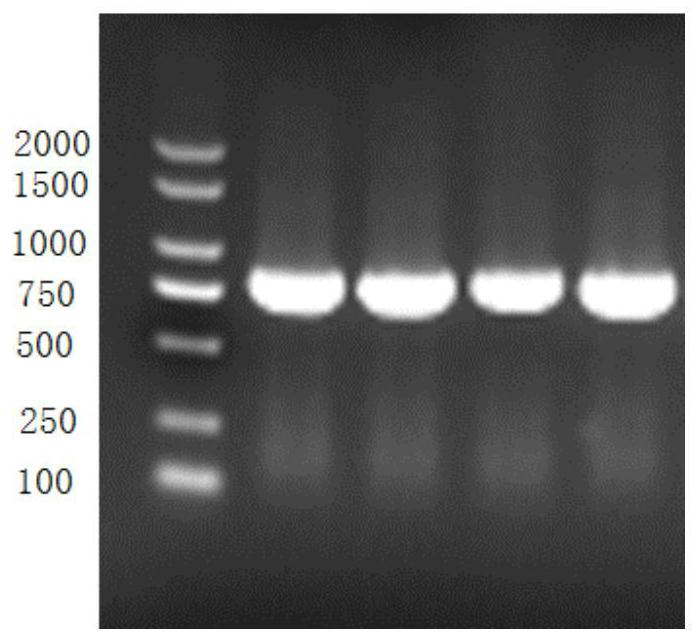
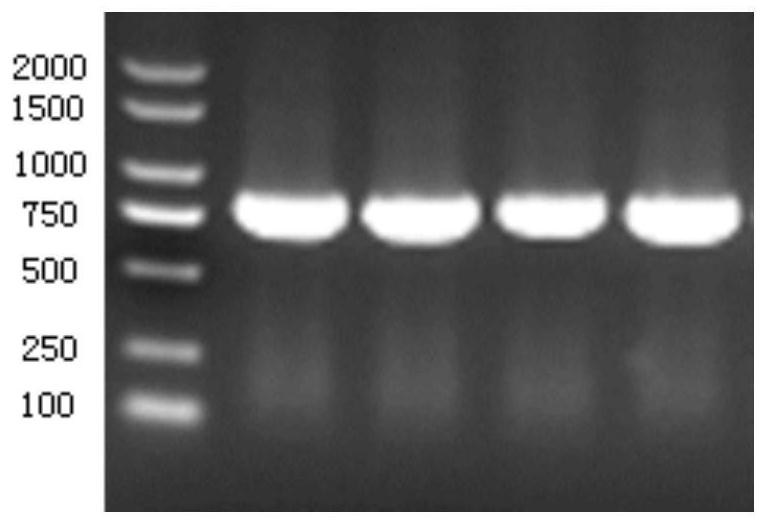
[0078] The obtained PCR product was analyzed and detected by 1% agarose gel electrophoresis, and an electrophoresis band with a size of about 711 bp was obtained. Transform the pET28a-LbudC plasmid into E.coli DH5α competent cells by electroporation, spread on solid LB solid medium containing 50 μg / mL kanamycin and culture overnight at 37°C, pick positive recombinants at -80°C Preserva...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com