Method for quantitatively detecting colletotrichum gloeosporioides by utilizing real-time fluorescent PCR (Polymerase Chain Reaction)
A real-time fluorescence quantification technology, applied in the biological field, can solve the problems of low detection sensitivity, economic loss, easy to miss the best time for prevention and control, etc., and achieve the effect of high specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1 A kind of method that utilizes real-time fluorescent PCR to quantitatively detect Colletotrichum spp.
[0054] 1). Design of specific primers
[0055] DNAMAN software was used to perform multiple sequence alignment of the conserved regions of ACT gene sequences of Glycoporum anthracis, and the conserved regions within Glycoporum anthracis species were selected, and specific primers ACT3F / ACT3R and ACT4F / ACT4R were designed using OLIGO7 software.
[0056] 2). Grind biological samples with liquid nitrogen, and then extract genomic DNA by CTAB method [Yan, L.; Zhang, C.; Ding, L.; Ma, Z. Development of a real-time PCR assay for the detection of Cladosporium fulvum in tomato leaves. J Appl Microbiol 2008, 104, 1417-1424, doi: 10.1111 / j.1365-2672.2007.03660.x].
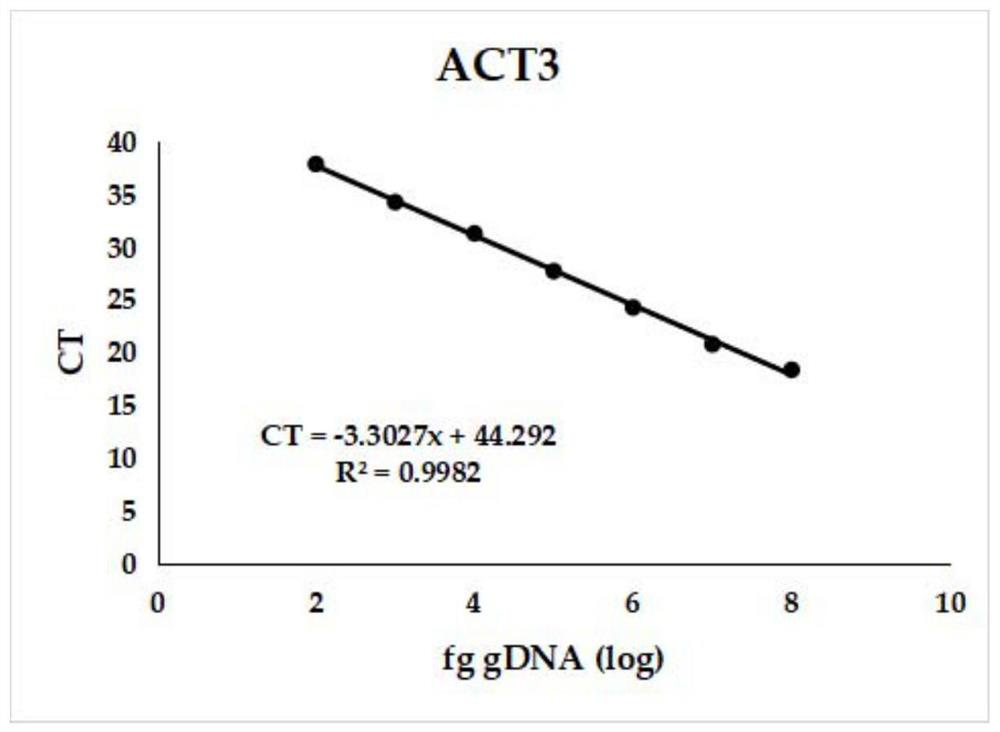
[0057] 3). Real-time fluorescent PCR to establish a linear regression curve
[0058] The obtained gDNA was subjected to ten-fold gradient dilution, and the diluted gDNA of different concentrations was...
Embodiment 2
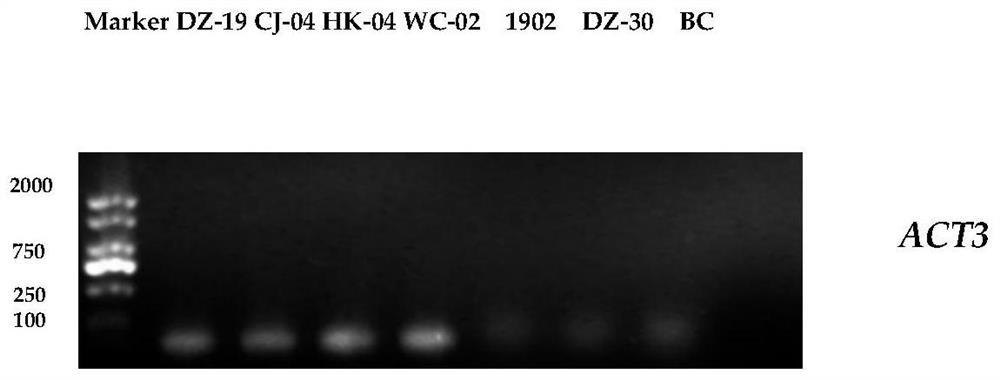
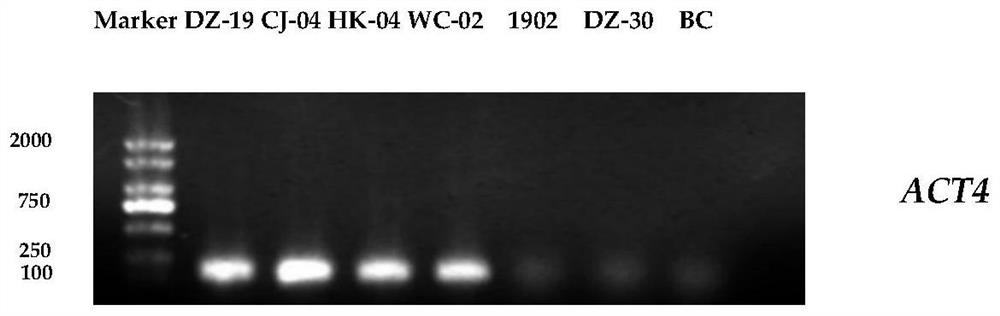
[0068] Example 2 Primer specificity test
[0069] (1) Using CTAB method to extract the DNA of Colletosporum anthracis, oxysporum anthracis, karst anthracis, Botrytis cinerea and the DNA of the plant material infected by gliasporum anthracis;
[0070] (2) Using 1% agarose gel electrophoresis, PCR amplification was carried out with the DNA of Colletotrichum, Anthracis oxysporum, Anthracis karstii, and Botrytis cinerea as templates, and ACT3F / ACT3R and ACT4F / ACT4R as primers. The amplified products were subjected to 1% agarose gel electrophoresis.
[0071] The PCR reaction system is:
[0072]
[0073] The reaction procedure is:
[0074]
[0075]
[0076] The results showed that: ACT3 and ACT4 only had amplified bands in G. anthracis, and there were no amplified bands in other strains.
Embodiment 3
[0077] Example 3 Sensitivity test
[0078] Use the gDNA, pDNA, and conidia DNA of Colletosporum anthracis to be diluted ten-fold respectively. The diluted gDNA, pDNA, and conidia DNA are used as templates, and primers are used to carry out real-time fluorescent PCR, and the amplification of real-time fluorescent quantitative PCR. The conditions are the same as those in Example 1, and a linear regression curve between the threshold cycle (ct) and the logarithm of the template concentration is established (see Figures 3 to 8 ).
[0079] The results showed that: ACT3 and ACT4 could detect at least 100 fg gDNA, 100 copies of pDNA, and 20 conidia DNA.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com