Rapid DNA sulfite conversion reagent and method
A sulfite and reagent technology, applied in the field of molecular biology, can solve the problems of reduced sensitivity of analytical techniques, long reaction time, inconvenience, etc., and achieve the effects of reduced DNA degradation, less DNA loss, and wide clinical applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1: Sulfate conversion reaction of cultured colon cancer HT29 cell DNA specimens
[0093] 1. Preparation of cellular genomic DNA
[0094] Colon cancer HT29 cell line was cultured according to conventional cell culture methods
[0095] Human monocyte-macrophage THP1 cell line as control cells
[0096] Take 0.5ml of cultured cells (cell density 1×10 7 ), using Qiagen Blood & Cell Culture DNAKits (Cat. 13323), according to the manufacturer's instructions, to extract cellular genomic DNA.
[0097] Take 0.1 μg of DNA for the following sulfite reaction.
[0098] 2. Sulfite conversion reaction system
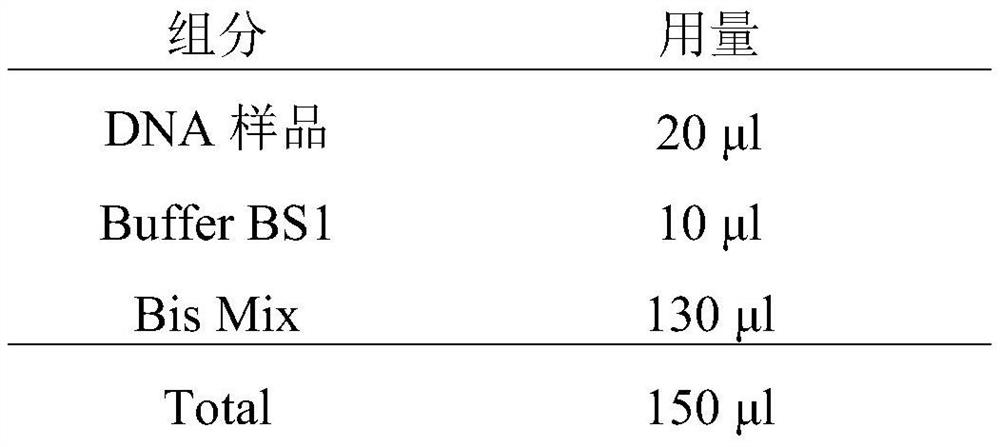
[0099] Prepare a sulfite conversion reaction system in a 0.5ml PCR tube. The specific preparation system is as follows:
[0100]
[0101] Buffer BS1: DNA protection solution (300mM hydroquinone aqueous solution)
[0102] Bis Mix: 10M Sulfite Conversion Solution (NaHSO 3 , (NH 4 ) 2 SO 3 , NH 4 HSO 3 , pH 5.2–5.3)
[0103] 3. Sulfite conversion reaction
...
Embodiment 2
[0106] Example 2: DNA desulfonation and purification recovery after sulfite treatment
[0107] The procedure according to the inventive method is as follows:
[0108] (1) After the sulfite treatment, transfer the reaction system in the tube to a clean 1.5ml centrifuge tube after a brief centrifugation, add 1ml of 6M guanidine hydrochloride and 40μl of magnetic beads, vortex and mix for 10sec, and let stand at room temperature for 5min;
[0109] (2) Place the centrifuge tube on the magnetic stand for 1 min, and discard the liquid after the magnetic beads are adsorbed;
[0110] (3) Add 600 μl of rinsing solution (containing 80% alcohol, 50 mM Tris Buffer), vortex and mix for 10 sec;
[0111] (4) Place the centrifuge tube on a magnetic stand for 1 min, and discard the liquid after the magnetic beads are adsorbed;
[0112] (5) Add 600 μl of desulfurization solution (containing 0.3N NaOH, 90% EtOH), vortex and mix for 10 sec, and leave at room temperature for 15 min;
[0113](6)...
Embodiment 3
[0122] Example 3: MSP detection of SDC2 methylation gene in HT29 intestinal cancer cells
[0123] SDC2 methylation gene analysis was performed on the transformed DNA of colon cancer cells with the following primers and probes:
[0124] SDC2_MF: TTAATAAGTGAGAGGGCGTCGC (SEQ ID NO: 1)
[0125] SDC2_MR: CGACTCAAACTCGAAAACTC (SEQ ID NO: 2)
[0126] SDC2 TaqMan probe: ROX-CGTAGTTGCGGGCGGCGGGAGTAGGC-BHQ2 (SEQ ID NO:3)
[0127] As an internal control, the transformed ACTB gene was detected by the following primers and probes
[0128] ACTB_MF: TGGTGATGGAGGAGGGTTTAGTAAGT (SEQ ID NO: 4)
[0129] ACTB_MR: AACCAATAAAACCTACTCCTCCCTTAA (SEQ ID NO: 5)
[0130] ACTB TaqMan probe: CY5-TGTGTTTGTTATTGTGTGTTGGGTGGTGGT-BHQ3 (SEQ ID NO:6)
[0131] Take 5 μl of purified transformed DNA for Real-time PCR analysis of human SDC2 gene methylation:
[0132] (1) Take 0.25μl of primers (SDC2_MF, SDC2_MR, ACTB_MF, ACTB_MR) and 0.125μl of fluorescent probes (SDC2 TaqMan probe, ACTB TaqMan probe), 6μl of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com