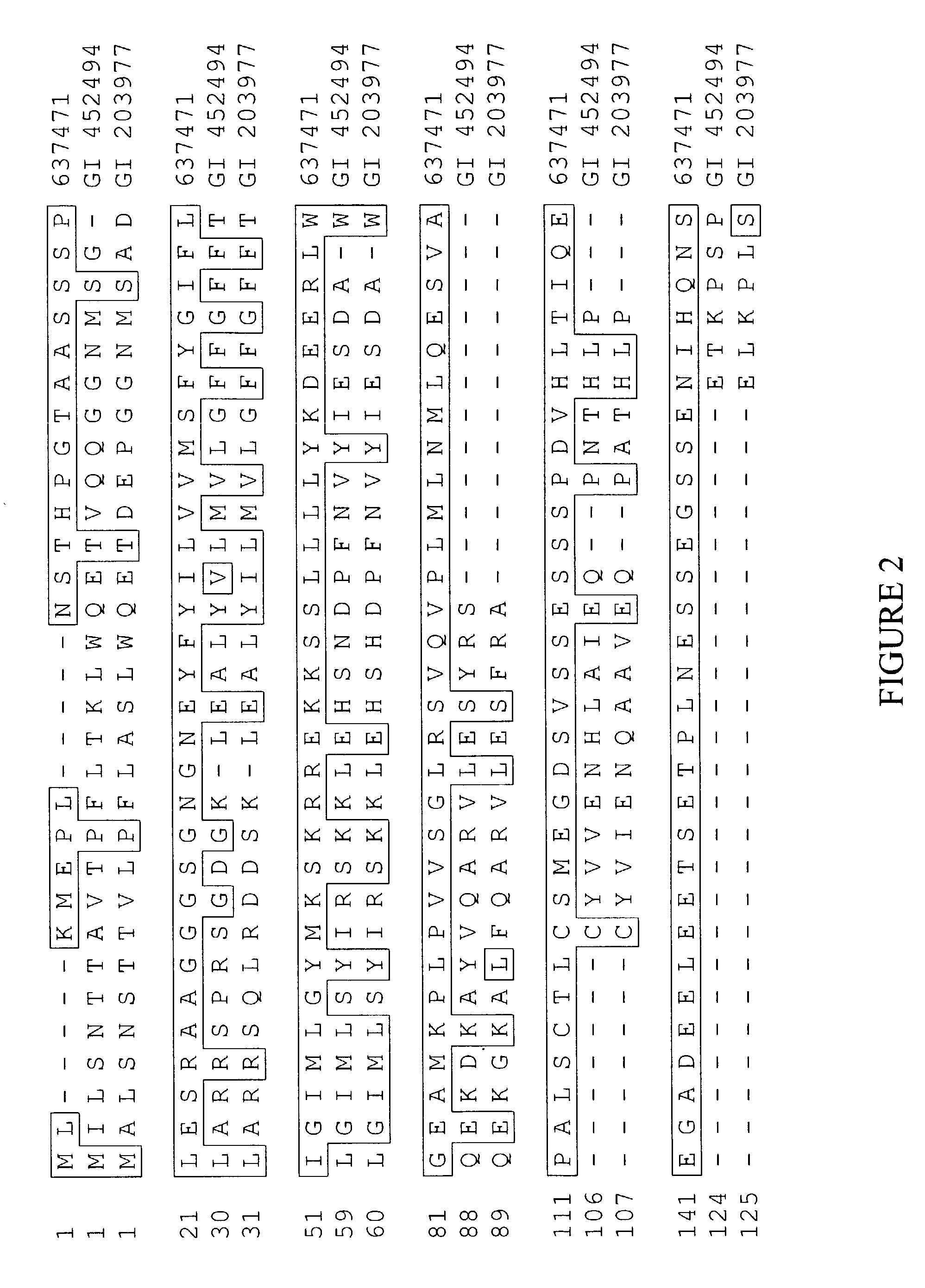
[0081] The
nucleic acid sequences encoding DRPCS may be extended utilizing a partial
nucleotide sequence and employing various PCR-based methods known in the art to detect upstream sequences, such as promoters and regulatory elements. For example, one method which may be employed, restriction-site PCR, uses universal and nested primers to amplify unknown sequence from
genomic DNA within a
cloning vector. (See, e.g., Sarkar, G. (1993) PCR Methods Applic. 2:318-322.) Another method, inverse PCR, uses primers that extend in divergent directions to amplify unknown sequence from a circularized template. The template is derived from restriction fragments comprising a known genomic locus and surrounding sequences. (See, e.g., Triglia, T. et al. (1988) Nucleic Acids Res. 16:8186.) A third method, capture PCR, involves PCR amplification of
DNA fragments adjacent to known sequences in human and
yeast artificial
chromosome DNA. (See, e.g., Lagerstrom, M. et al. (1991) PCR Methods Applic. 1:111-119.) In this method, multiple
restriction enzyme digestions and ligations may be used to insert an engineered double-stranded sequence into a region of unknown sequence before performing PCR. Other methods which may be used to retrieve unknown sequences are known in the art. (See, e.g., Parker, J. D. et al. (1991) Nucleic Acids Res. 19:3055-3060). Additionally, one may use PCR, nested primers, and PROMOTERFINDER libraries to walk
genomic DNA (Clontech, Palo Alto, Calif.). This procedure avoids the need to screen libraries and is useful in finding
intron /
exon junctions. For all PCR-based methods, primers may be designed using commercially available
software, such as OLIGO 4.06 primer
analysis software (National Biosciences Inc., Plymouth, Minn.) or another appropriate program, to be about 22 to 30 nucleotides in length, to have a
GC content of about 50% or more, and to anneal to the template at temperatures of about 68.degree. C. to 72.degree. C.
[0088] In order to express a biologically active DRPCS, the
nucleotide sequences encoding DRPCS or derivatives thereof may be inserted into an appropriate
expression vector, i.e., a vector which contains the necessary elements for transcriptional and translational control of the inserted coding sequence in a suitable host. These elements include regulatory sequences, such as enhancers, constitutive and
inducible promoters, and 5' and 3' untranslated regions in the vector and in
polynucleotide sequences encoding DRPCS. Such elements may vary in their strength and specificity. Specific
initiation signals may also be used to achieve more efficient translation of sequences encoding DRPCS. Such signals include the ATG
initiation codon and adjacent sequences, e.g. the Kozak sequence. In cases where sequences encoding DRPCS and its
initiation codon and upstream regulatory sequences are inserted into the appropriate
expression vector, no additional transcriptional or translational control signals may be needed. However, in cases where only coding sequence, or a fragment thereof, is inserted, exogenous translational control signals including an in-frame ATG initiation codon should be provided by the vector. Exogenous translational elements and initiation codons may be of various origins, both natural and synthetic. The efficiency of expression may be enhanced by the inclusion of enhancers appropriate for the particular host
cell system used. (See, e.g., Scharf, D. et al. (1994) Results Probl.
Cell Differ. 20:125-162.)
[0096] For long term production of recombinant proteins in mammalian systems, stable expression of DRPCS in
cell lines is preferred. For example, sequences encoding DRPCS can be transformed into
cell lines using expression vectors which may contain viral origins of replication and / or endogenous expression elements and a
selectable marker gene on the same or on a separate vector. Following the introduction of the vector, cells may be allowed to grow for about 1 to 2 days in enriched media before being switched to selective media. The purpose of the
selectable marker is to confer resistance to a selective agent, and its presence allows growth and
recovery of cells which successfully express the introduced sequences. Resistant clones of stably transformed cells may be propagated using
tissue culture techniques appropriate to the
cell type.
[0116] In other embodiments, any of the proteins, antagonists, antibodies, agonists, complementary sequences, or vectors of the invention may be administered in combination with other appropriate therapeutic agents. Selection of the appropriate agents for use in
combination therapy may be made by one of ordinary skill in the art, according to conventional pharmaceutical principles. The combination of therapeutic agents may act synergistically to effect the treatment or prevention of the various disorders described above. Using this approach, one may be able to achieve therapeutic
efficacy with lower dosages of each agent, thus reducing the potential for adverse side effects.
[0129] Ribozymes, enzymatic
RNA molecules, may also be used to catalyze the specific cleavage of
RNA. The mechanism of
ribozyme action involves sequence-specific hybridization of the
ribozyme molecule to complementary target
RNA, followed by endonucleolytic cleavage. For example, engineered hammerhead motif
ribozyme molecules may specifically and efficiently catalyze endonucleolytic cleavage of sequences encoding DRPCS.
[0162] With respect to
cancer, the presence of a relatively high amount of transcript in biopsied tissue from an individual may indicate a predisposition for the development of the
disease, or may provide a means for detecting the
disease prior to the appearance of actual clinical symptoms. A more definitive diagnosis of this type may allow
health professionals to employ preventative measures or aggressive treatment earlier thereby preventing the development or further progression of the
cancer.
 Login to View More
Login to View More