Assembly and screening of highly complex and fully human antibody repertoire in yeast
a highly complex and human antibody technology, applied in the field of generating libraries of recombinant expression vectors, can solve the problems of long breeding period of transgenic mice, inability to select high affinity antibodies, and inability to readily process, assemble, or express/secrete functional antibodies,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
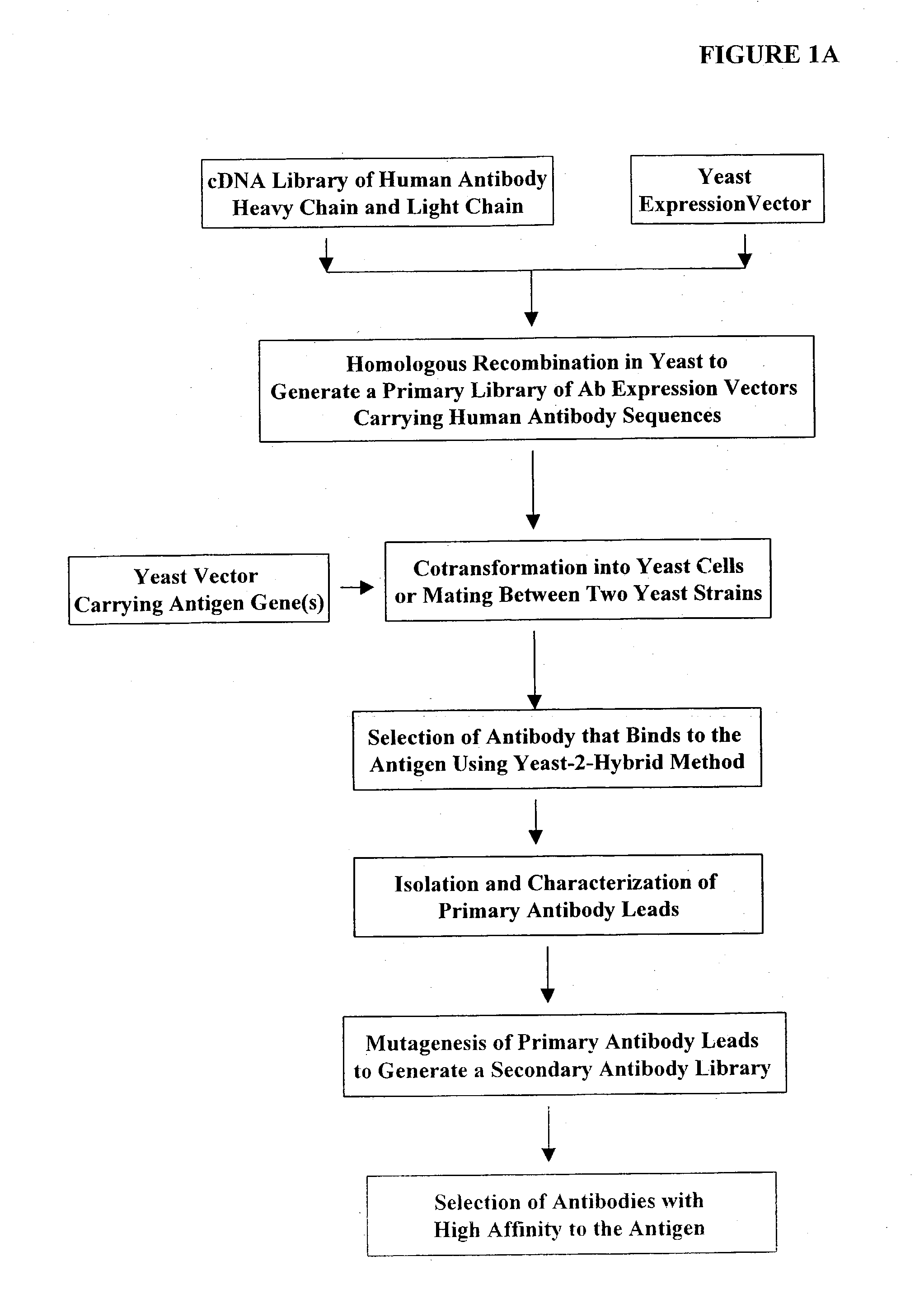
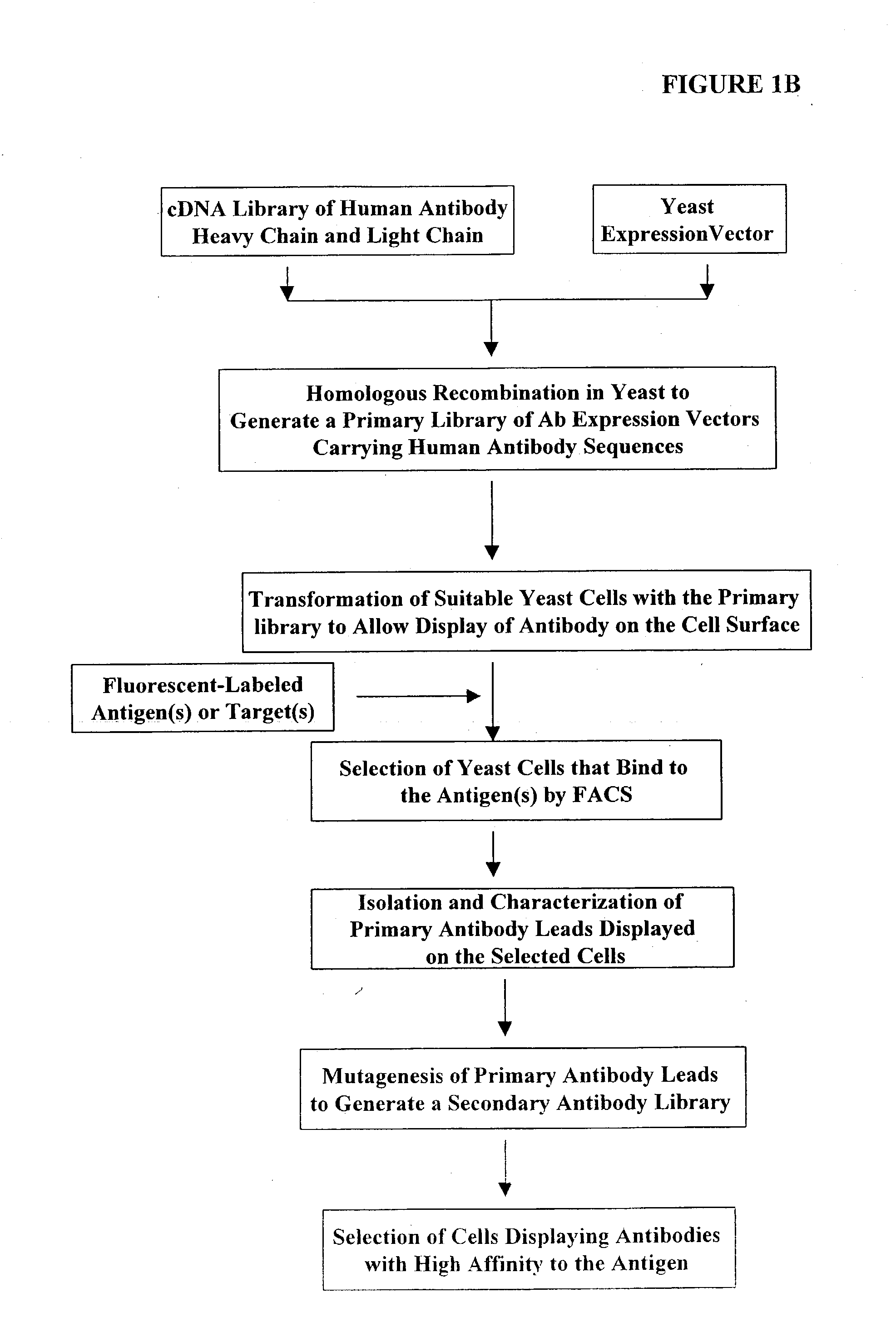
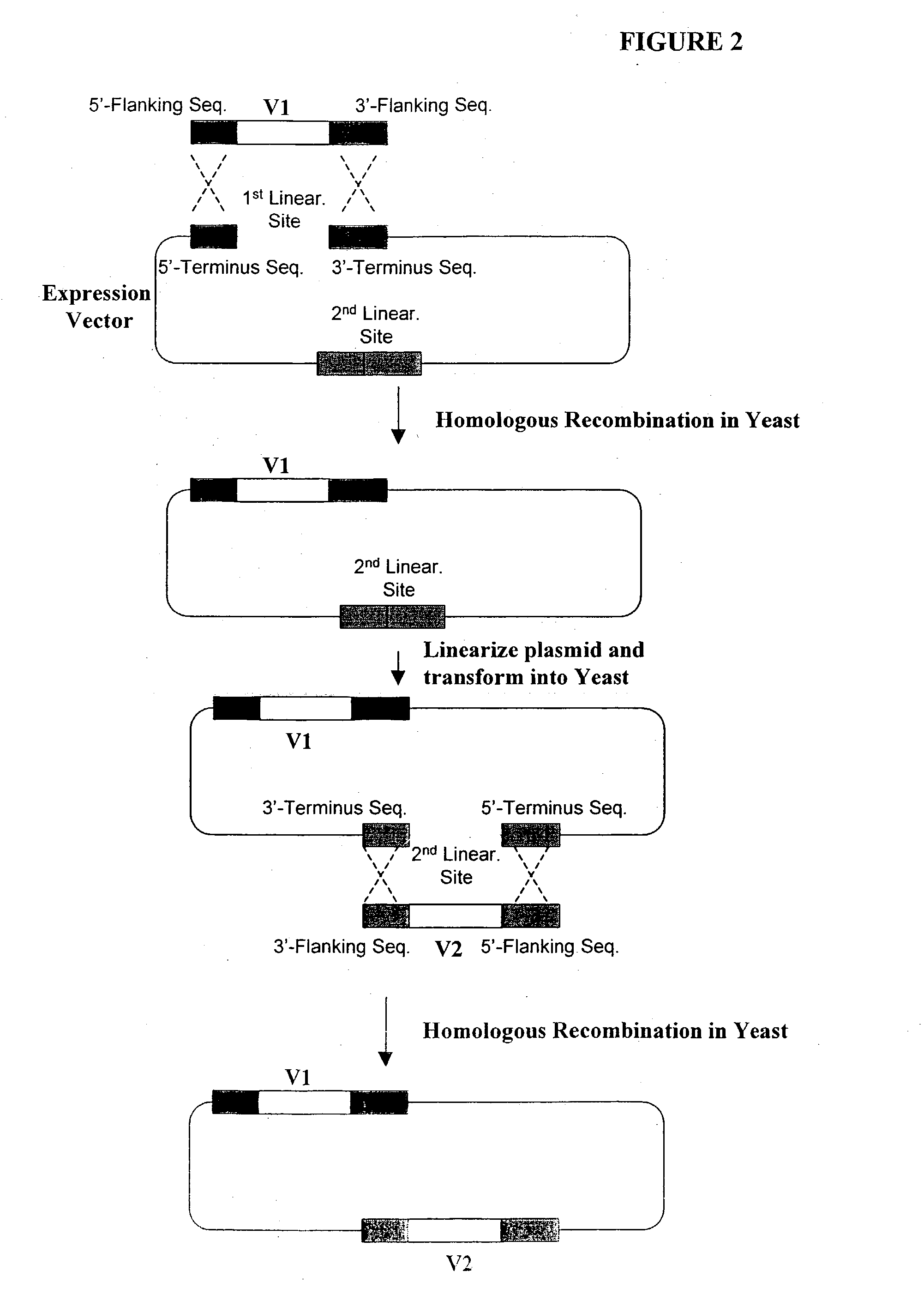
[0391] Construction of Expression Vectors Containing Human Antibody Library Using Homologous Recombination in Vivo
[0392] The following illustrates examples of how to use general homologous recombination as an efficient way of constructing recombinant human antibody library. The coding sequence of each member of the antibody library includes heavy-chain and light chain regions derived from a library of human antibody repertoire. The light chain region of the antibody is fused with a two-hybrid system activation domain (AD) to form a two-hybrid expression vector in the yeast. In an alternative design, the light chain region of the antibody is fused with Aga2 subunit of yeast a-agglutinin to form a surface dislay expression vector in the yeast. The heavy chain region of the antibody is expressed separately from the light chain region by a different promoter.
[0393] 1) Isolation of Human Antibody cDNA Gene Pool
[0394] A complex human antibody cDNA gene pool is generated by using the metho...
example 2
[0432] Screening of Antibody Libraries in Yeast with the Two-Hybrid System against Defined Protein Antigens via Mating between Two Yeast Strains
[0433] This example describes a procedure used to screen the antibody libraries generated in the Example 1. The human antibody libraries are generated in yeast strain with an .alpha. mating type. This mating type of yeast can be readily mated with an a type of yeast with simple mating procedure to form diploid yeast cells. Guthrie and Fink (1991) "Guide to yeast genetics and molecular biology" in Methods in Enzymology (Academic Press, San Diego) 194:1-932. The a-yeast contains the target (probe, or bait) plasmid.
[0434] The target plasmid contains a fusion formed between the GAL 4 DNA binding domain (BD) and any desired target protein that is to be used as a probe to fish out the antibodies as its affinity ligand. When the two types of yeast cell mate and form diploid cells, the probe plasmid and the library clone plasmid also come together i...
example 3
[0437] Screening of Human Antibody Libraries against a Library of Antigens in a Yeast Two-Hybrid System.
[0438] For small number of pre-selected probes (i.e. baits or targets), the procedure of individual mating screening as described above is sufficient. However, this procedure can also be modified to suit for screening against large number of probes. The following list describes the potential probes that are in large number and may not suitable for individual mating screening:
[0439] a. A collection of human EST clones, or total library of human EST. Such EST collection can be ordered from public resource in a library format with individually clones arrayed in 96-well or 384-well plates. The EST inserts from the original collection (usually in bacterial cloning and sequencing vectors) are PCR amplified with extended homologous sequences at both ends. The EST inserts can be PCR amplified and additional flanking sequences can be added to both ends of the ESTs by PCR for mediating homo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| dissociation constant | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com