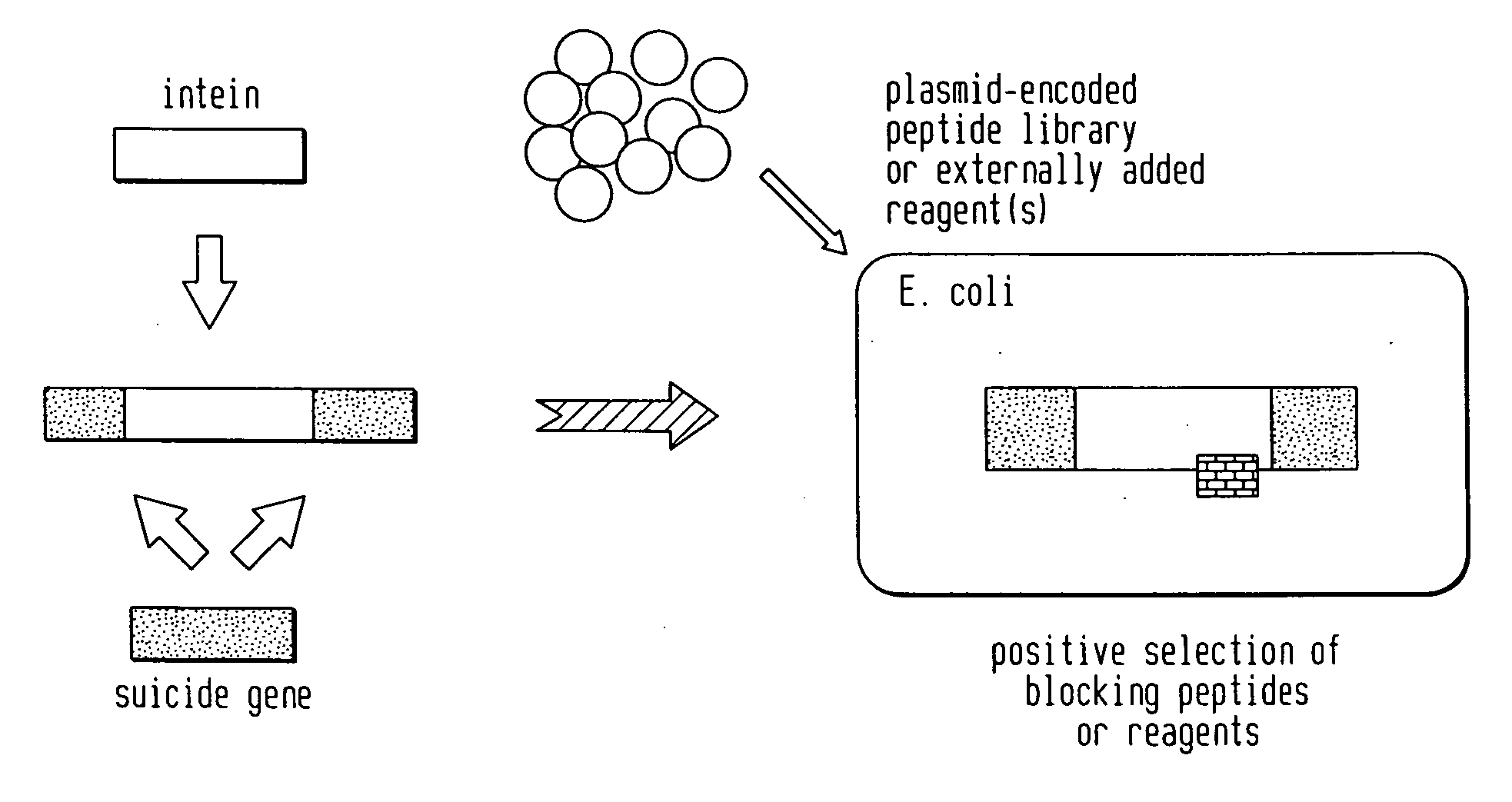
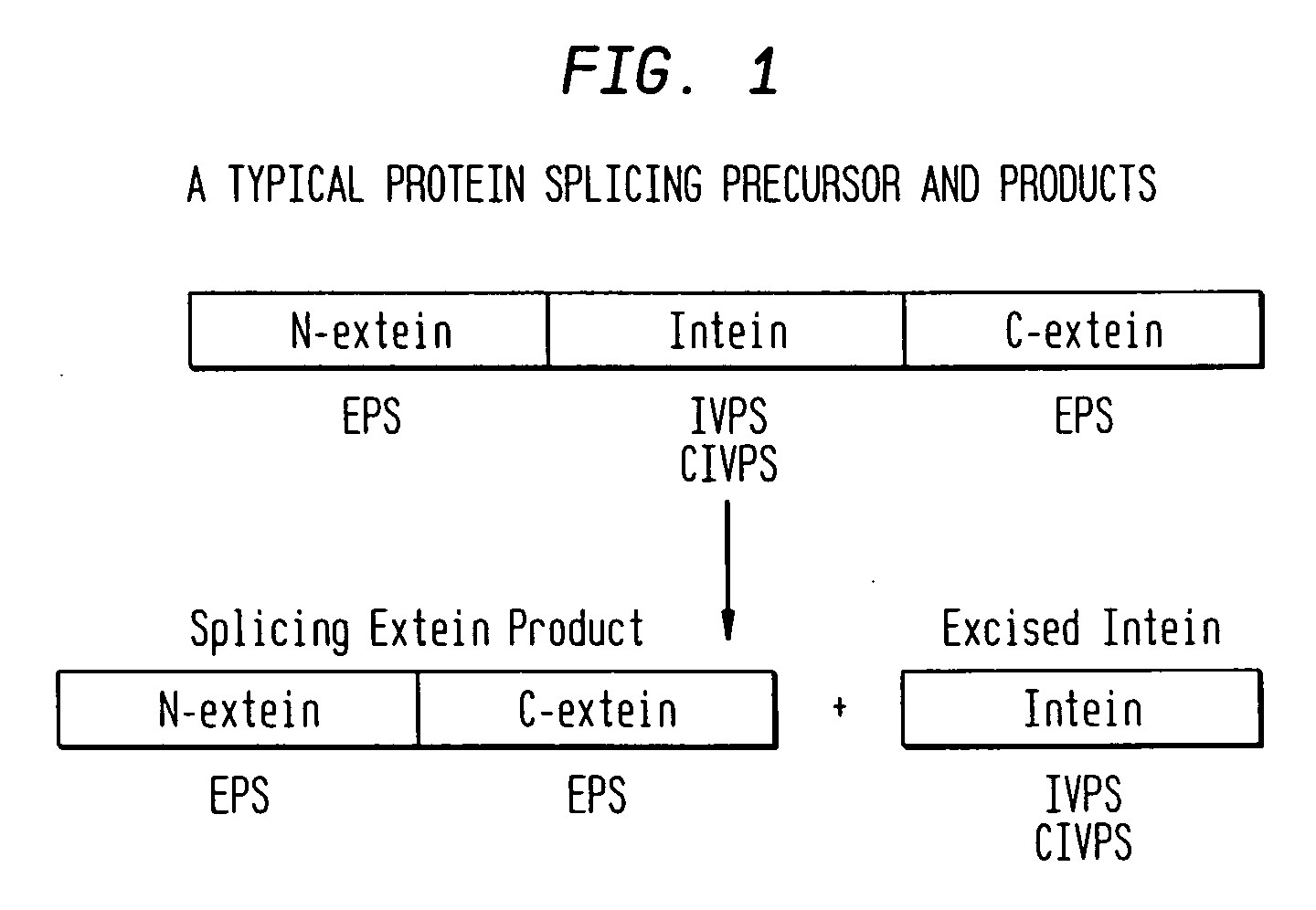
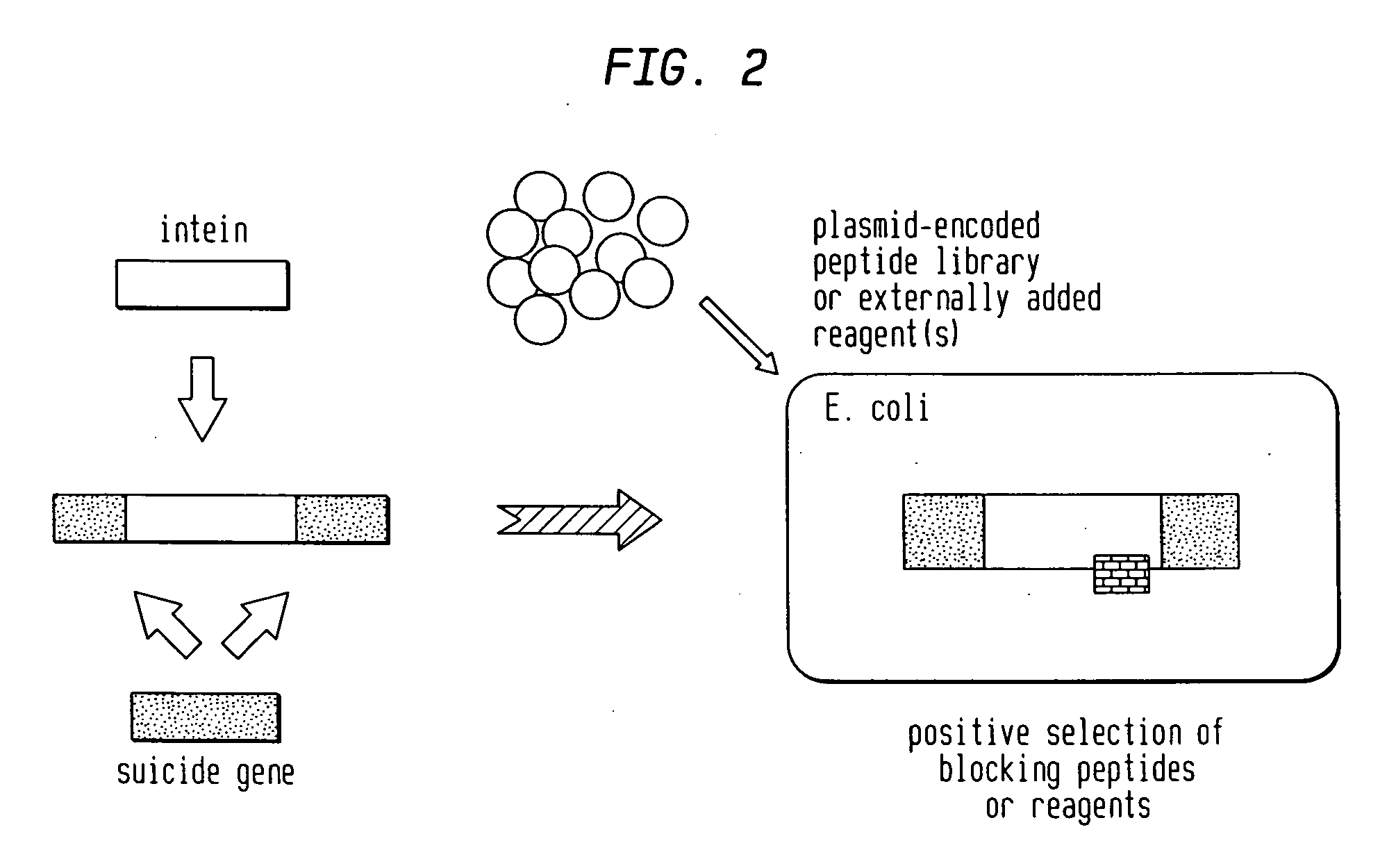
Screening and use of reagents which block or activate intein splicing utilizing natural or homologous exteins
a technology of intein splicing and reagents, applied in the direction of antibacterial agents, polypeptides with his-tags, instruments, etc., can solve the problems of inefficient protein splicing in the foreign extein context, foreign extein sequences may not be acceptable substrates at all, and the system suffers from several limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
A Mxe GyrA Intein-Mediated Positive Selection System for Inhibition of Protein Splicing
A Positive Selection System Based on Blocking Splicing of the Mxe GyrA Intein (IVPS) in E. coli GyrA: Background
[0064] Gyrases are essential multimeric enzymes involved in DNA replication in bacteria (Swanberg and Wang, 1987). Both gyrase subunit A and B have been extensively studied as drug targets in bacterial human pathogens (e.g., Mycobacteria, Salmonella, Enterbacteriaceae, Citrobacter, Pseudomonas, Streptococcus, Staphylococcus, Yersinia, Rhodobacter, Haemophilus, Neisseria, Providencia). The GyrA subunit of gyrases can complex with quinoline drugs, such as ofloxacin, and induce cell death. The complex formation of quinolines with gyrase is followed by a rapid and irreversible inhibition of DNA synthesis, inhibition of growth, and induction of the SOS response (see FIG. 3A). At higher drug concentrations, cell death occurs as double-strand DNA breaks in the bacterial chromosome are releas...
example ii
A M. Tuberculosis DnaB Intein-Mediated Positive Selection System
A Positive Selection System Using the Mtu DnaB Intein (IVPS) in its Native Mtu DnaB Extein in E. coli: Background
[0083] The hexameric E. coli helicase encoded by the dnaB gene interacts with an hexameric DnaC complex and ATP. Some DnaB mutants are dominant lethal (Bouvier and Oreglia, C.R. Acad. Sci. Hebd. Seances Acad. Sci. D., 280:649-652 (1975), Maurer and Wong, J. Bacteriol 170:3682-3688 (1988), Saluja and Godson, J. Bacteriol. 177:1104-1111 (1995) and Sclafani, et al., Mol. Gen. Genet., 182:112-118 (1981)). By dominant or dominantly cytotoxic, we mean that the toxicity occurs even if homologous proteins are present which are not cytotoxic or resistant to the drug, i.e., the cytotoxic effect dominates irrespective of the presence of non-cytotoxic homologs. The mutant protein is deficient in ATP hydrolysis, helicase activity, and replication activity at the chromosomal origin of replication resulting in cell death...
example iii
In Vivo Control of Protein Splicing for Chemotherapeutic Purposes or to Make Controllable Gene Knockouts
[0098] The selection and screening systems described for selection of agents that modulate protein splicing can also be applied to intein-less versions of the extein gene to select for agents that inhibit or activate the extein gene product. All of the selection and screening systems described in this patent are based on the activity or inactivity of the extein portion of the precursor. If one deletes the intein from the intein-containing gene by methods known to one skilled in the art, then one can select for agents that block or activate extein activity also using the methods described for inhibiting or activating splicing of the intein containing precursor, since these latter methods involve assaying extein function. For example, if one deletes the intein from the Mtu DnaB gene by methods known to one skilled in the art, then one can select for agents that block activity of th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com