Production of Biologically Active Proteins
a technology of applied in the field of biologically active recombinant peptides and proteins, can solve the problems of poor host for the synthesis of correctly-folded mammalian proteins normally stabilized by disulfide bonds, lack of biological activity expected of that material, and poor efficiency of the oxi-redox system of bacteria for eukaryotic proteins, etc., to achieve the effect of increasing the accumulation rate and being readily availabl
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Accumulation of RX3-ECFP Derived Fusion Proteins in Dense Fractions of Transfected Mammal Cells
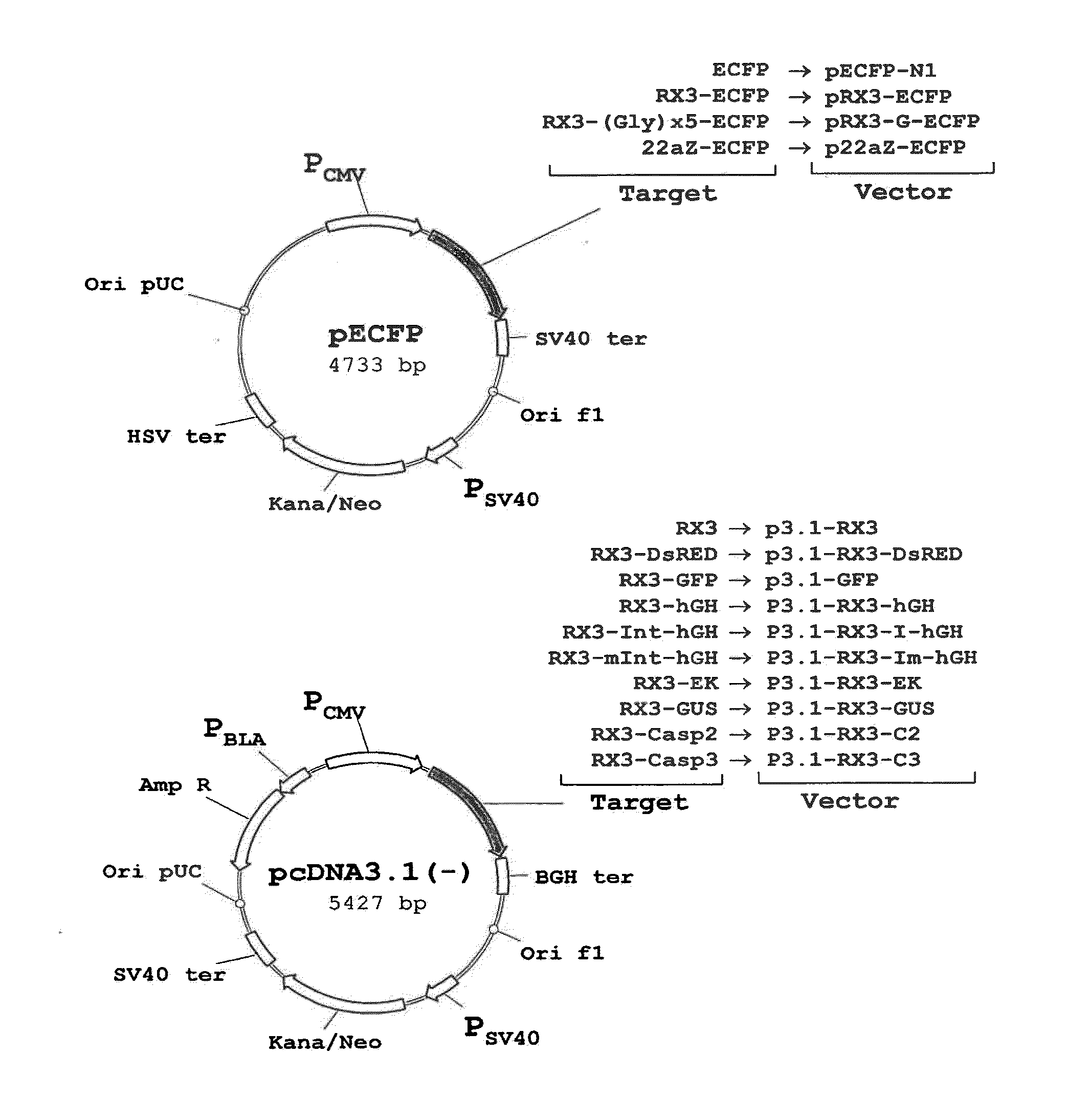
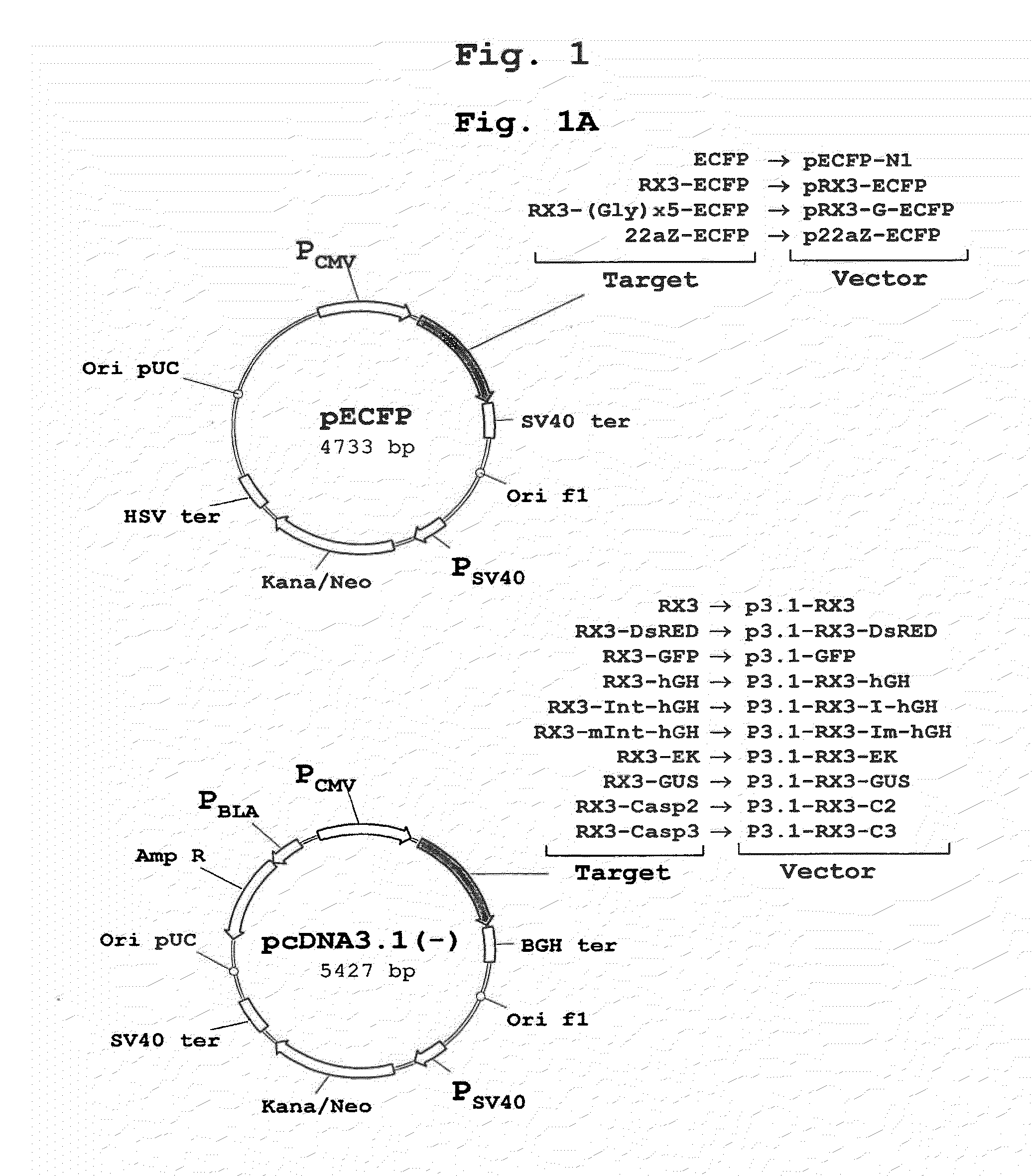
[0172]The polynucleotide sequence coding for the N-terminal gamma-zein coding sequence RX3 (WO2004003207) was fused directly, or through a linker consisting of five glycines, to the 5′ end of the sequence encoding ECFP, a cyan fluorescent variant of GFP. Those constructs (FIG. 1A) that code for the fusion proteins RX3-ECFP or RX3-Gx5-ECFP were introduced in cultured mammalian CHO cells by the Lipofectamine-based transfection method (Invitrogen). CHO cells transfected with plasmid pECFP-N1 (Clontech) containing the gene sequence of a non-targeted (cytosolic) ECFP were used as controls.
[0173]Transfected mammalian cell extracts were loaded on density step gradients and centrifuged. The accumulation of recombinant proteins in the different fractions was analyzed by immunoblot. The results shown in FIG. 2A indicate that RX3-ECFP and RX3-Gx5-ECFP sedimented in fractions F42, F56 and P correspond...
example 2
Accumulation of Active ECFP Fused to PBIS Domains in RPBLAs of Transfected Mammal Cells
[0175]To determine if the fusion proteins RX3-ECFP, RX3-Gx5-ECFP and 22aZ-ECFP are active inside the RPBLAs, confocal microscopic analysis was performed to visualize target protein fluorescence in transfected CHO cells (FIG. 1A). Cyan fluorescence was imaged by excitation at 458 nm with an argon ion laser with an emission window set at 470-530 nm. As shown in FIG. 3, the corresponding fusion proteins, RX3-ECFP (FIG. 3A) and RX3-Gx5-ECFP (FIG. 3B) and 22aZ-ECFP (FIG. 3D), were detected in proximity to the ER, indicating that the gamma-zein and the alpha-zein signal peptides are functional in mammalian cells where they mediates the translocation of the fusion protein into the ER.
[0176]In addition, the fusion proteins surprisingly also accumulated preferentially into large and dense spherical structures that strongly resembled both authentic PBs of cereal seed and RPBLAs in heterologous systems visua...
example 3
Subcellular Localization of Other Fluorescent Proteins Fused to RX3 in CHO Cells
[0178]The sub-cellular localization of RX3-DsRED and RX3-GFP fusion proteins in transiently transfected CHO cells was analyzed by confocal microscopy to analyze whether other fluorescent proteins than ECFP fused to RX3 are properly folded and bioactive inside RPBLAs. It is important to note that DsRED shares no homology to ECFP, which implies a completely different folding mechanism. Fluorescence images from the transfected cells were obtained by using a confocal laser scanning microscope (Leica TCS SP, Heidelberg, Germany) fitted with spectrophotometers for emission band wavelength selection. Green fluorescent images were collected by excitation at 488 nm with an Argon ion laser using an emission window set at 495-535 nm. Red fluorescent images were collected using 543 nm excitation with a HeNe laser and an emission window of 550-600 nm. Optical sections were 0.5 μm thick.
[0179]The expression of RX3-GFP...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameters | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com