Methods for reprogramming cells and uses thereof
a cell reprogramming and cell technology, applied in the field of eukaryotic cell reprogramming, can solve the problems of revealing true reprogramming, difficult control, wrong cell type, etc., and achieve the effects of reducing ethical and time constraints, reducing the cost of cell therapy, and being more potent and/or safer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
[0277]The examples set forth herein below provide exemplary methods for obtaining Reprogrammed and Dedifferentiated cells, including Neural Stem-Like Cells (NSLCs). Also provided are exemplary protocols, molecular tools, probes, primers and techniques.
example i
Preparation of Human Fibroblast Cells
[0278]Human Foreskin fibroblast (HFF) cells were purchased from American Type Culture Collection (ATCC, Manassas, Va.) and expanded in cell culture flasks with Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen), supplemented with 10% heat-inactivated fetal calf serum (FCS, Hyclone Laboratories), 0.1 mM non-essential amino acids, and 1.0 mM sodium pyruvate (invitrogen) at 37° C., 5% CO2. The medium was changed twice per week. Cells were trypsinized using Trypsin 0.25% for 4 minutes at 37° C., followed by adding trypsin inhibitor solution, pelleting the cells by centrifugation, washing the cells once with PBS, and plating the cells at a ratio of 1:2 onto tissue culture flasks until a suitable number of cells was reached.
[0279]Cells were then trypsinized and plated (8×104 cells / well) in cell culture plates pre-coated with Laminin (10 μg / ml, Sigma) in two different composition of CDM medium: CDM I Medium consisting of a 3:1 ratio of Dulbecco's mod...
example ii
Comparison of Reprogramming Efficiency of Three Different Neurogenic Genes
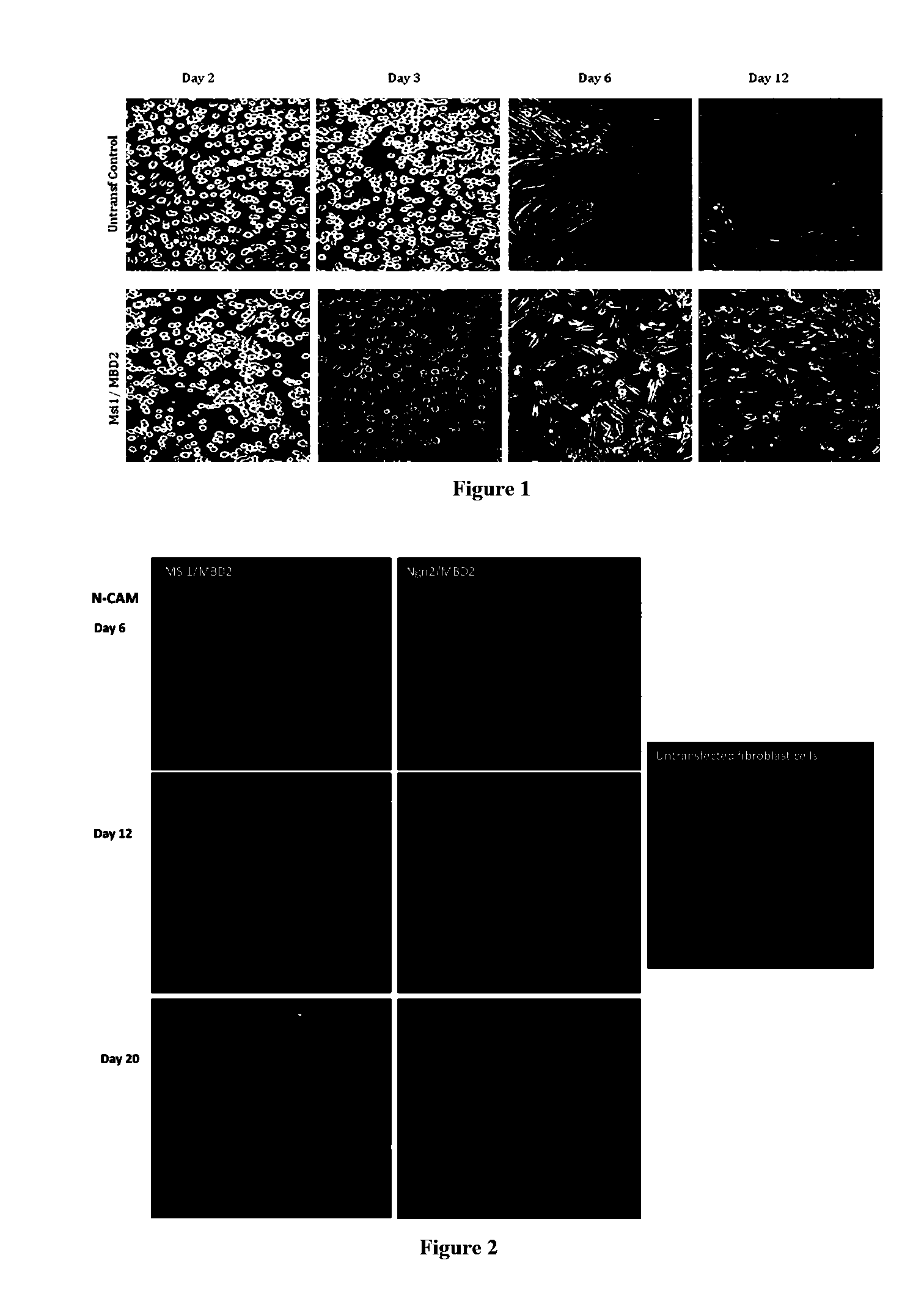
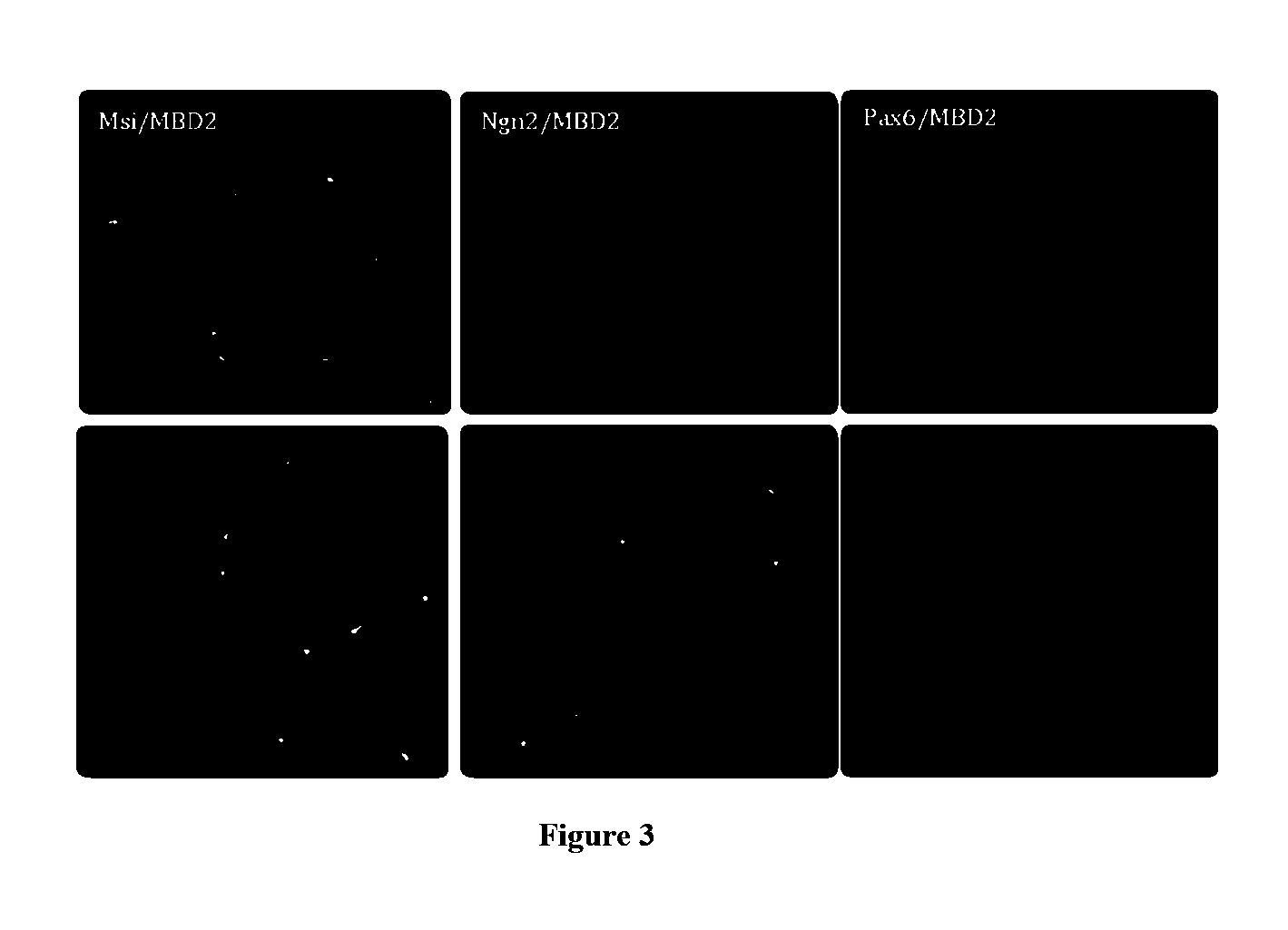
[0292]HFF cells were cultured as described in Example I and plated in CDM I medium. Cells were transfected using the Amaxa Nucleofector™ Device (Lanza). The HFFs were harvested with TrypLE™ (Gibco), resuspended in CDM Medium and centrifuged for 10 min at 90×g (1×106 cells / tube). The supernatant was discarded and gently resuspended in 100 μl of Basic Nucleofectar™ Solution (basic Nucleofector™ kit for primary mammalian fibroblasts, Lanza). Each 100 μl of cell suspension was combined with a different mix of plasmid DNA (for example, sample 1 was mixed with 2 μg of pCMV6-XL5-Pax6 and 2 μg pCMV6-XL5-MBD2). Cell suspension was transferred into an Amaxa certified cuvette and transfected with the appropriate program (U023). The sample was transferred without any further resuspension into a coated culture plate with LAS-Lysine / Alanine (BrainBits™, 50 μg / ml) and the cells were incubated at 37° C., 5% CO2. These steps w...
PUM
| Property | Measurement | Unit |
|---|---|---|
| three-dimensional structure | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| compositions | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com