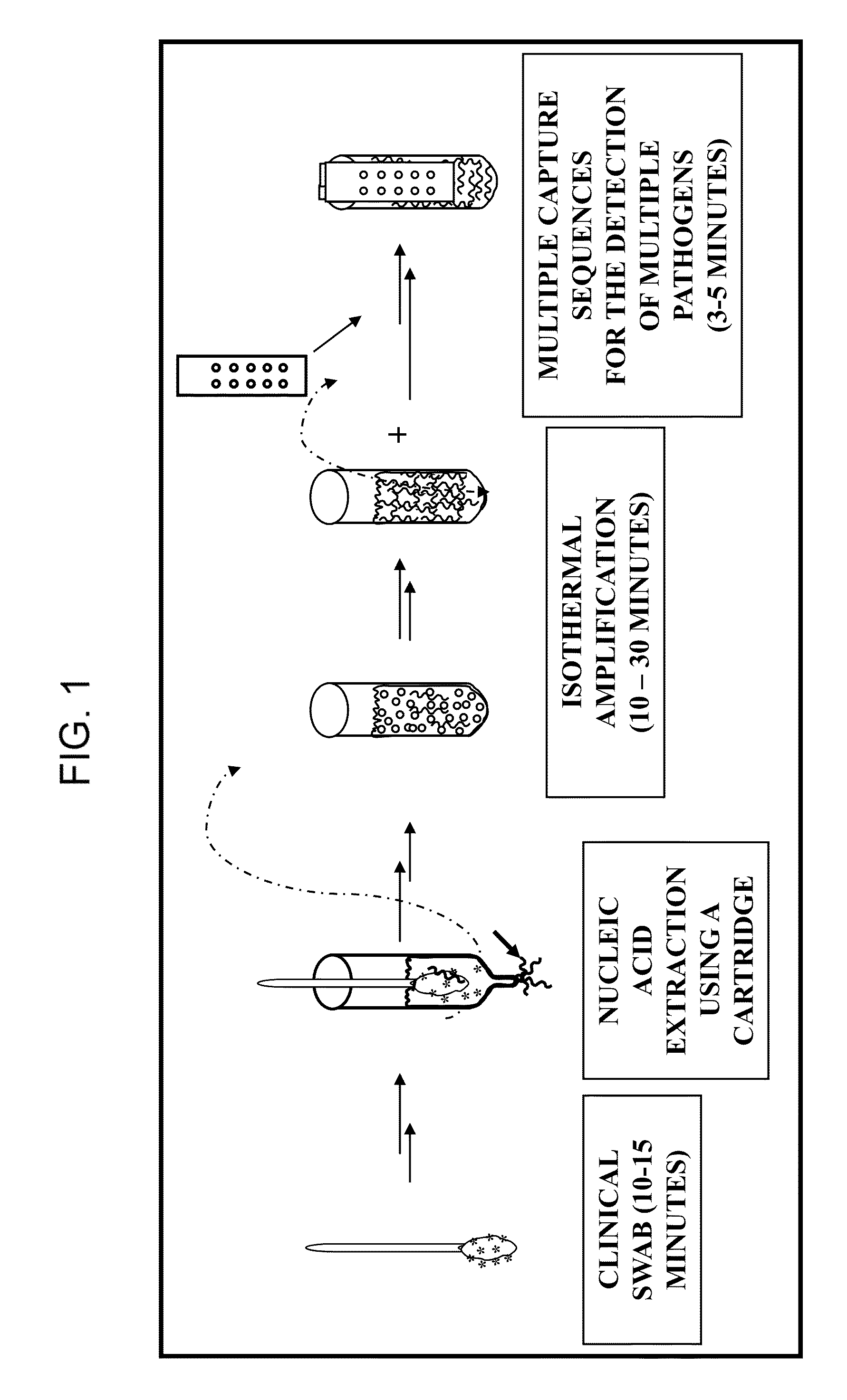
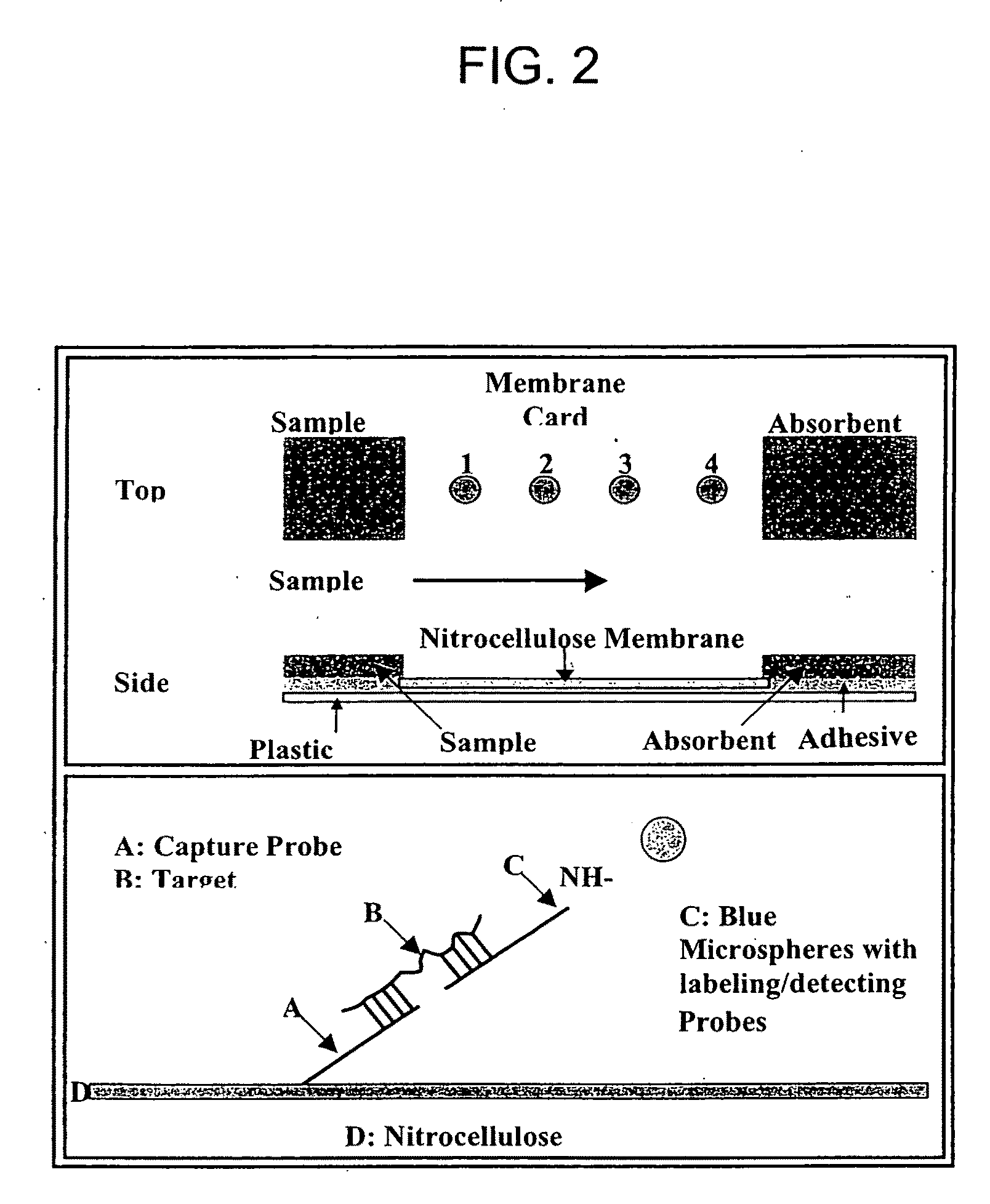
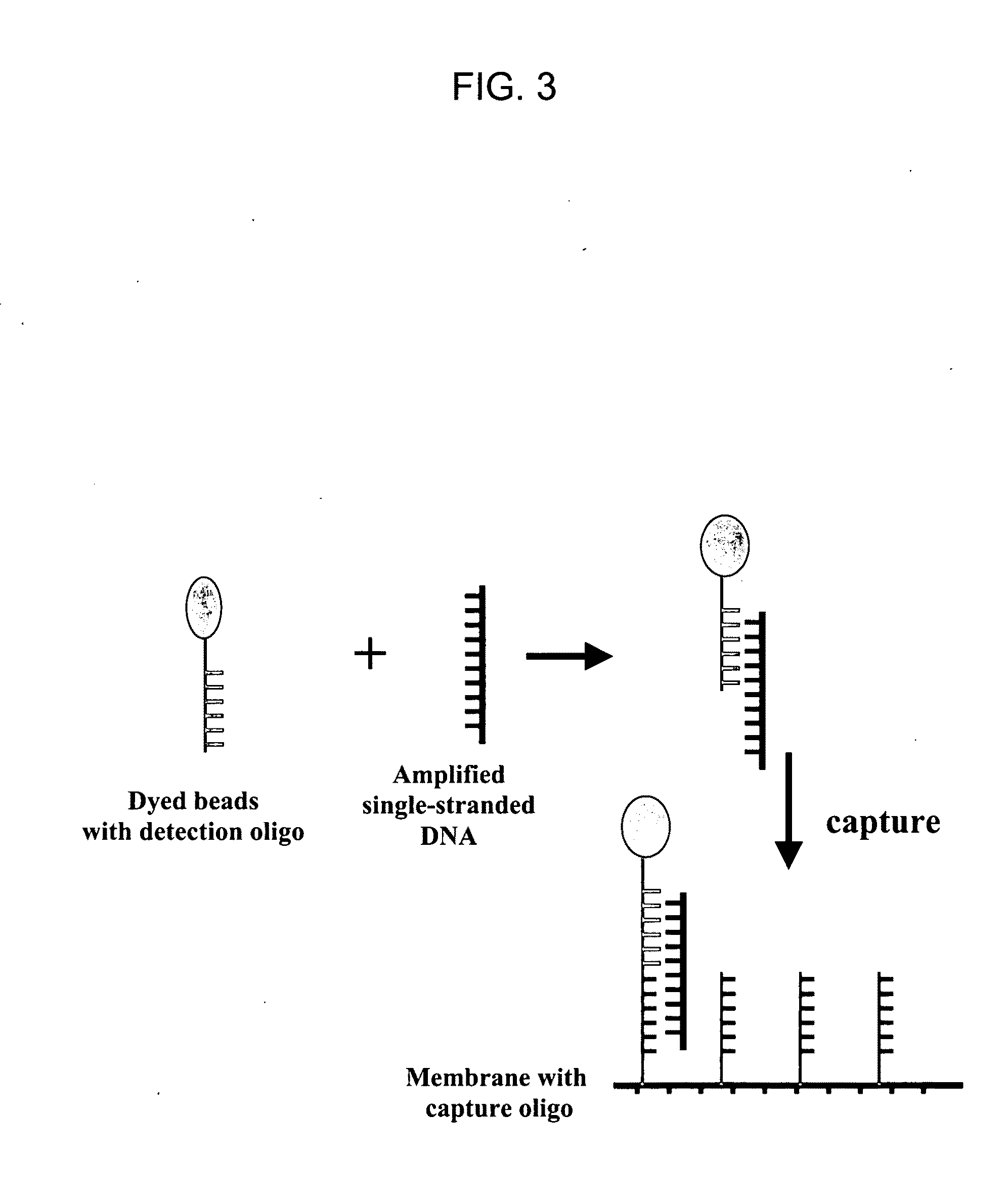
Nucleic Acid Detection System and Method for Detecting Influenza
a detection system and technology for nucleic acids, applied in combinational chemistry, biochemistry apparatus and processes, library screening, etc., can solve the problems of global pandemic of influenza, significant economic loss, social panic, and localized epidemics, and achieve unprecedented assay speed and simplicity, and minimize the extent
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Amplification and Detection of Bacillus Target DNA Using HDA and Lateral Flow Capture
[0119]This example evaluates the performance of HDA in combination with the lateral flow detection platform of the invention, using Bacillus anthracis (Ba) as a model target, and demonstrates the feasibility, sensitivity and specificity of these component of the detection system of the invention applied to bacterial DNA targets.
Materials and Methods:
[0120]Conjugation of Labeling / Detecting Oligonucleotide Probes onto Dyed Microspheres:
[0121]Carboxyl-polystyrene microspheres embedded with blue dyes, with diameters from 0.08-0.39 μM, were purchased from Spherotech Inc. (Libertyville, Ill.). To label / detect target sequence (amplification product or synthetic target template oligomer), specific labeling / detecting probes carrying an amine modification group at their 5′ end (complementary to the target sequence) were covalently conjugated to the carboxylated microspheres using a standard EDAC (1-ethyl-3-(3...
example 2
Extraction and Amplification of Influenza RNA from Immunomagnetically Captured Virus
[0139]This example shows the effective extraction of amplifiable RNA from influenza A virus using a combination of immunomagnetic affinity capture of influenza A virus and NaOH lysis. Extracted RNA was subjected to RT-PCR as an indicator of the quality of the RNA so extracted.
Materials and Methods:
[0140]Influenza A strain, A / Sydney / 5 / 97 (H3N2 strain). Negative control, MDCK cells (not infected with virus). Positive control, viral RNA extracted from partially purified A / Sydney / 4 / 97 using Qiagen kit RNeasy (˜106 pfu).
[0141]For the first set of experiments, the bead-avidin / biotin-antiflu antibody system was used to isolate virus from media, isolated virion preparation was stored at −70° C. Frozen virion preparation was purified by filtration through a 0.2 μm filter post-infection of MDCK cells, so the samples contained a significant amount of cell debris. No other handling of the virus was performed pri...
example 3
Amplification of Influenza RNA Using Isothermal Rt-HDA and Detection Using Lateral Flow Nucleic Acid Assay
[0147]This example demonstrates successful amplification and detection of influenza A viral RNA using isothermal RT-HDA in combination with lateral flow detection.
Materials and Methods:
[0148]Single-step RT-HDA reaction: A single reaction in a volume of 20 μl containing 10× HDA annealing buffer in IsoAmp® tHDA kit (Biohelix®, Beverly, Mass.), 100 nM influenza A-specific primers, influenza A RNA, 500 mM dNTPmix (Invitrogen, Carlsbad, Calif.), 5 mM MgCl2, 0.01M DTT, 40 units of RNaseOUT (Invitrogen, Carlsbad, Calif.), and 10 units of rBst DNA polymerase, Large Fragment (IsoTherm™) (EPICENTRE, Madison, Wis., USA), was prepared and incubated at 65° C. for 10 minutes. Then, 5 units of Hybridase (Thermostable RNaseH) (EPICENTRE, Madison, Wis., USA), was added to the reaction followed by 20 minutes incubation at 65° C. The HDA reaction was initiated by adding 10× HDA annealing buffer in...
PUM
| Property | Measurement | Unit |
|---|---|---|
| reaction volumes | aaaaa | aaaaa |
| reaction volumes | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com