Formaldehyde dehydrogenase genes from methylotrophic yeasts
a technology of methylotrophic yeast and dehydrogenase, which is applied in the field of methylotrophic yeast, can solve the problems of inconvenient monitoring of methanol concentration, insufficient promotion or convenience in all settings, and induction of psub>aox1/sub>-controlled expression strains in large-volume high-density fermentor cultures, etc., and achieves the effect of reducing the risk of toxicity and reducing the risk o
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of Formaldehyde Dehydrogenase-defective Mutants of P. pastoris
[0095]As a first step in cloning the P. pastoris formaldehyde dehydrogenase gene (FLD1), mutants were sought that were specifically defective in FLD activity. Previous biochemical studies of methylotrophic yeasts indicated that FLD is involved in the metabolism of both methanol as carbon source and methylamine as nitrogen source (Zwart et al., 1983). To search for P. pastoris fld1 mutants, nitrosoguanidine-mutagenized cultures were screened for strains that were unable to utilize methanol as carbon source and methylamine as nitrogen source. Complementation analysis and other classical genetic techniques were performed as described in Cregg and Russell (1998). Five mutants belonging to a single complementation group were identified.
[0096]These five strains were further examined by measuring the levels of activity of key methanol pathway enzymes in extracts prepared from methanol-induced cultures of each strain. ...
example 2
Isolation and Characterization of the P. pastoris FLD1 Gene
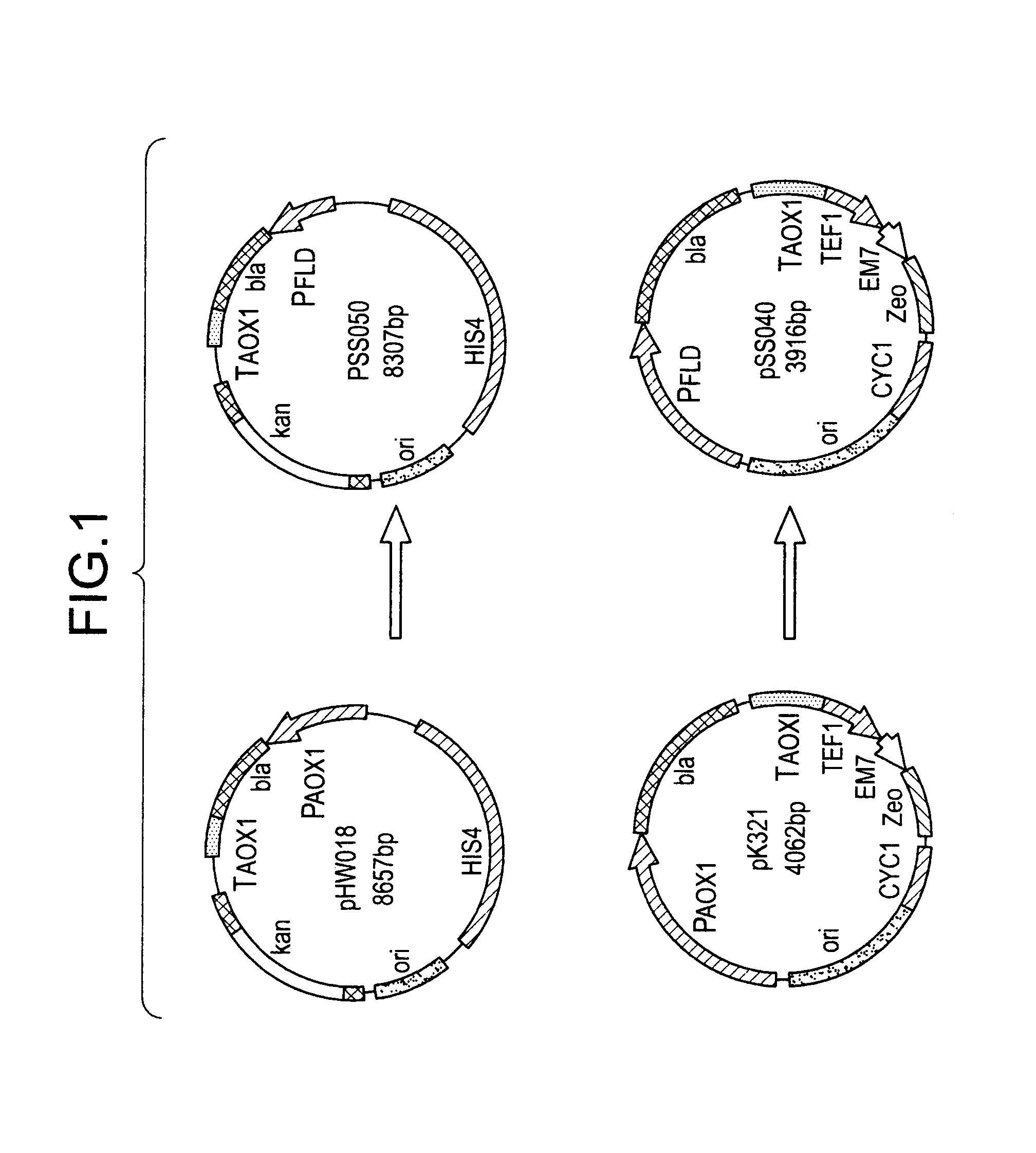
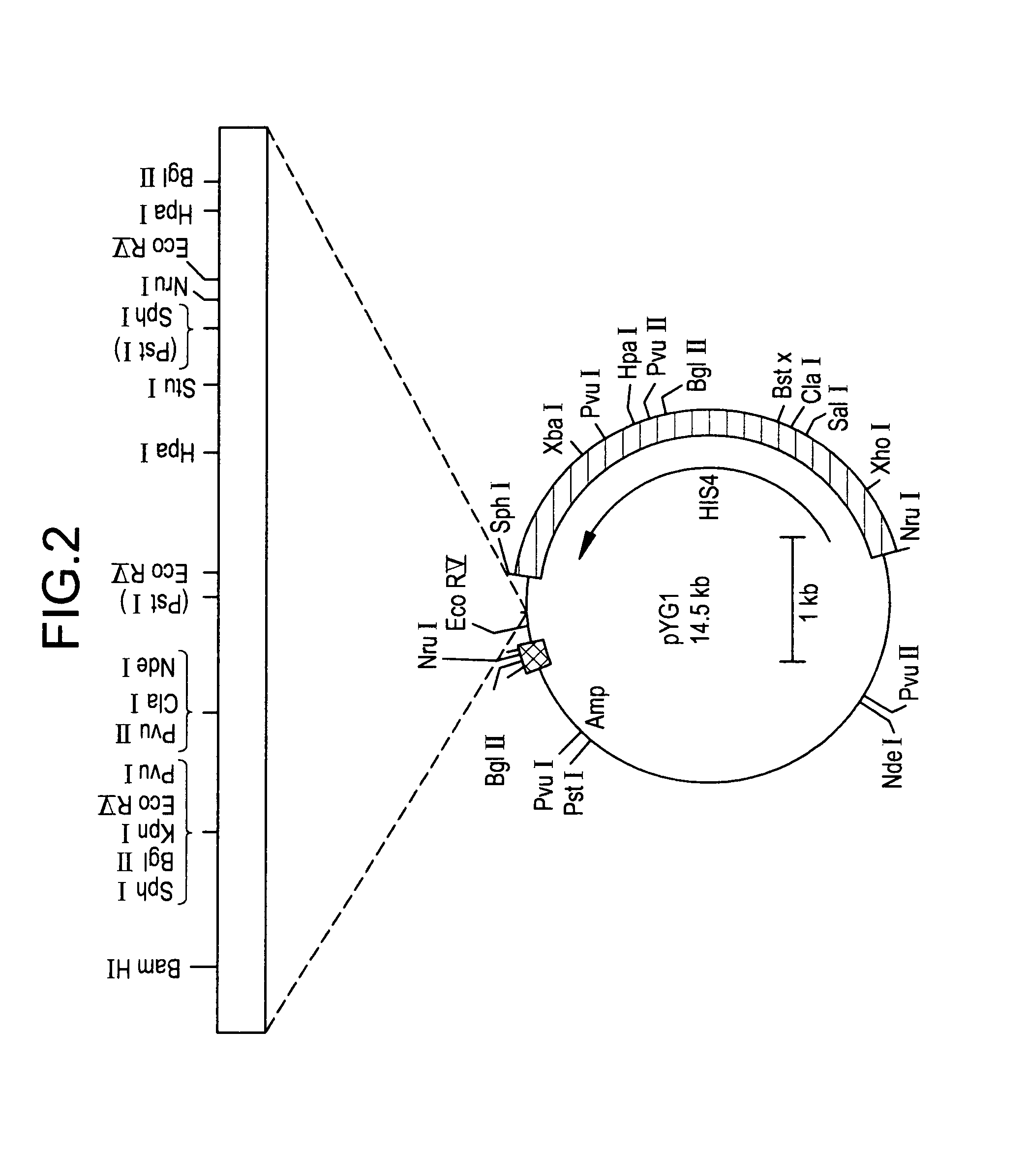
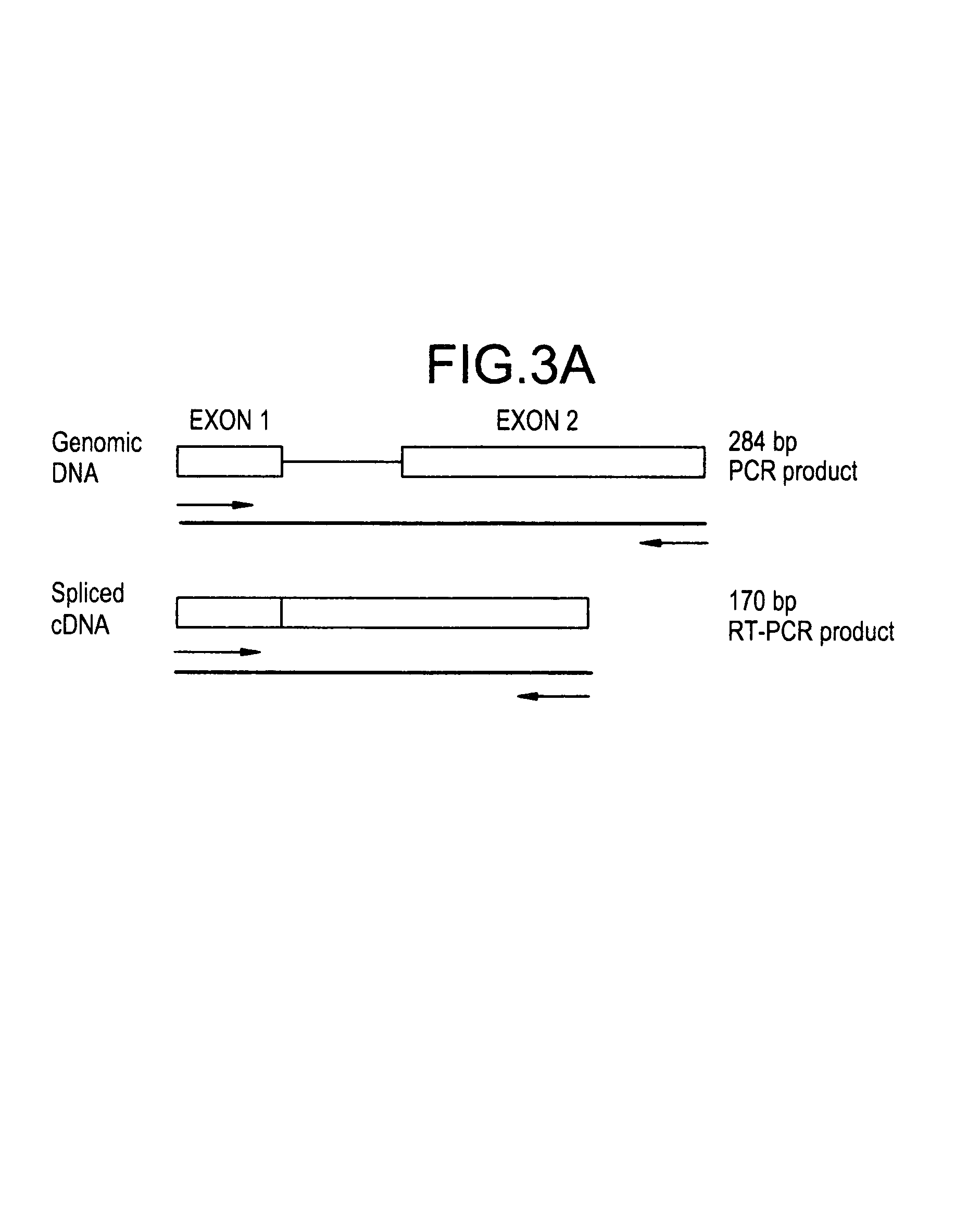
[0099]To clone the putative FLD1 gene by functional complementation, strain GS241 was first crossed to P. pastoris strain GS115 (his4) to obtain a derivative that was both methanol-utilization defective (Mut−) and auxotrophic for histidine (His−). One Mut−His− strain that resulted from this cross, MS105 (fld1-1 his4), was then transformed with 5–10 μg of a P. pastoris genomic DNA library constructed in the P. pastoris-E. coli shuttle vector pYM8 using the spheroplast method (Cregg et al., 1985; Liu et al., 1995). The plasmid pYM8 is composed of the Saccharomyces cerevisiae histidinol dehydrogenase gene (SHIS4) and a P. pastoris-specific autonomous replication sequence (PARS1) inserted into E. coli plasmid pBR322. Approximately 50,000 library transformants were selected for His+ prototrophy on YND medium agar and resultant selected clones further selected on YNM plates for Mut+ phenotype. Total DNA was extracted from a pool o...
example 3
Isolation and Characterization of the Hansenula polymorpha FLD Gene
[0105]The putative H. polymorpha FLD1 gene was isolated using the same functional complementation strategy described above for the P. pastoris gene. An H. polymorpha genomic DNA library was constructed in P. pastoris vector pYM8 in the same manner as the P. pastoris library (Liu et al. 1995). Briefly, H. polymorpha genomic DNA was partially digested with Sau3A and size selected for fragments of 5–20 kb. These fragments were ligated into the BamHI site of pYM8. The library was composed of approximately 100,000 independent E. coli transformants with greater than 90% containing an insert. The average size of insert DNA was approximately 10 kb. Assuming that the size of the H. polymorpha genome is 10,000 kb, the library contained approximately 100 genome equivalents of H. polymorpha genomic DNA. Plasmids were recovered and analyzed for ones that were capable of simultaneously retransforming MS105 (fld1-1 his4 to both His...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Electrical resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com