Method for producing western blot resin with dual recognition group polymer chain and application thereof
A western blotting and dual recognition technology, applied in the field of separation and enrichment of natural trace proteins, can solve the problems of single imprinted protein method, limit the practical significance of the application of western blotting polymers, etc. The effect of improving specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
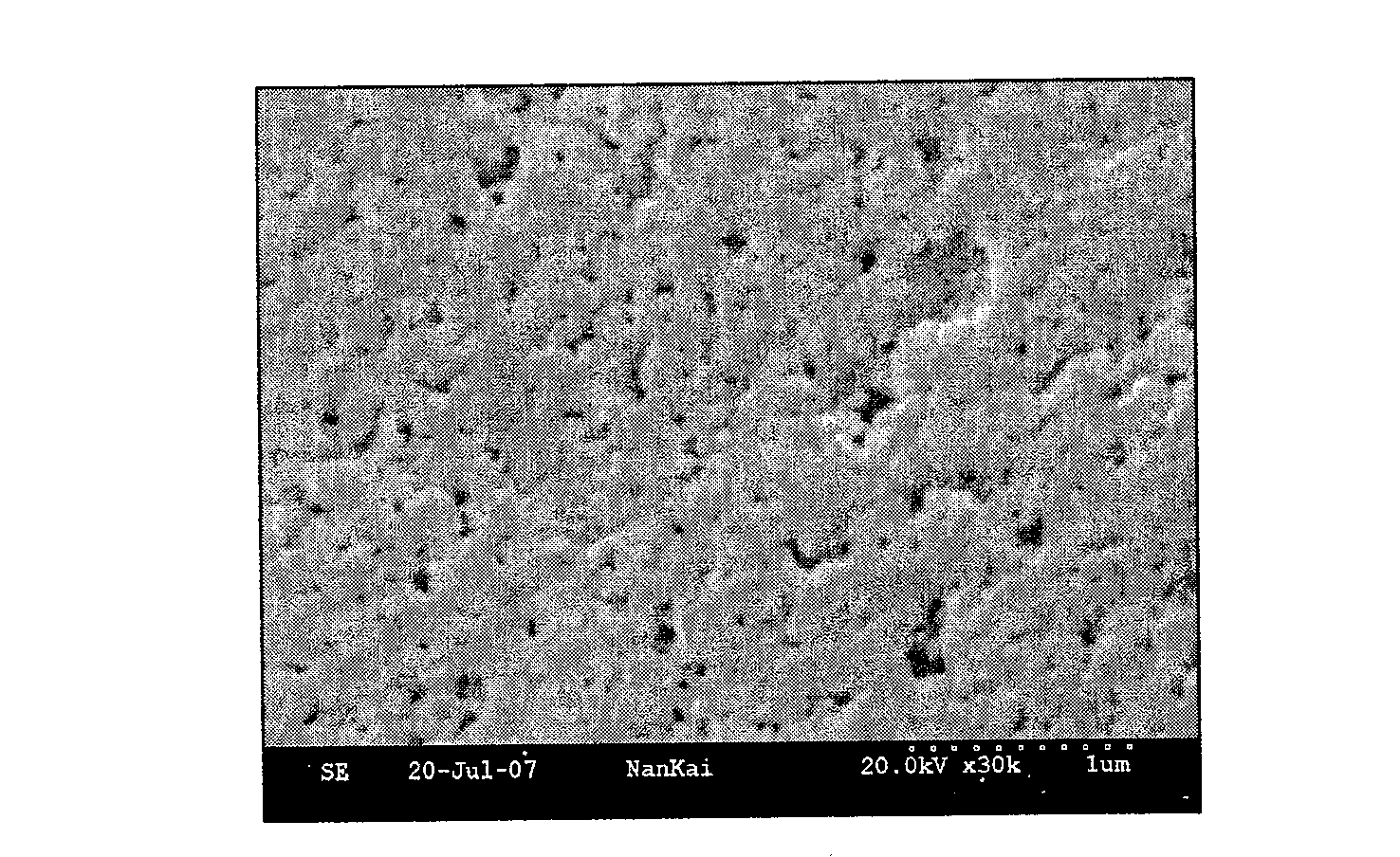
[0024] First, the preparation of the auxiliary recognition chain: take 2.3g of azobisisobutyronitrile (AIBN) as the initiator, 0.5ml of 4-vinylpyridine and 30ml of acrylonitrile (AN) as the reaction monomer, and 200ml of absolute ethanol as the solvent , and sequentially added to a pre-dried three-necked round-bottomed flask, and the molar ratio of acrylonitrile and 4-vinylpyridine was 1:99. The mass ratio of the reaction monomer acrylonitrile and the initiator azobisisobutyronitrile is 10.6:1. Nitrogen protection, electromagnetic stirring, 80 ° C water bath heating, reflux condensation, reaction 8hr. The product was poured into absolute ethanol to precipitate while it was hot, washed repeatedly, and dried in vacuum at 50° C. for 24 hours to obtain the random copolymer PANVP. Take 2 g of the refined random copolymer of acrylonitrile and 4-vinylpyridine (PANVP), add it to 15 ml of 85% (w / w) concentrated sulfuric acid, and hydrolyze it at 100° C. for 4 hours, so that the nitril...
Embodiment 2
[0027] Example 2, Application of Western Blot Resin in Protein Separation
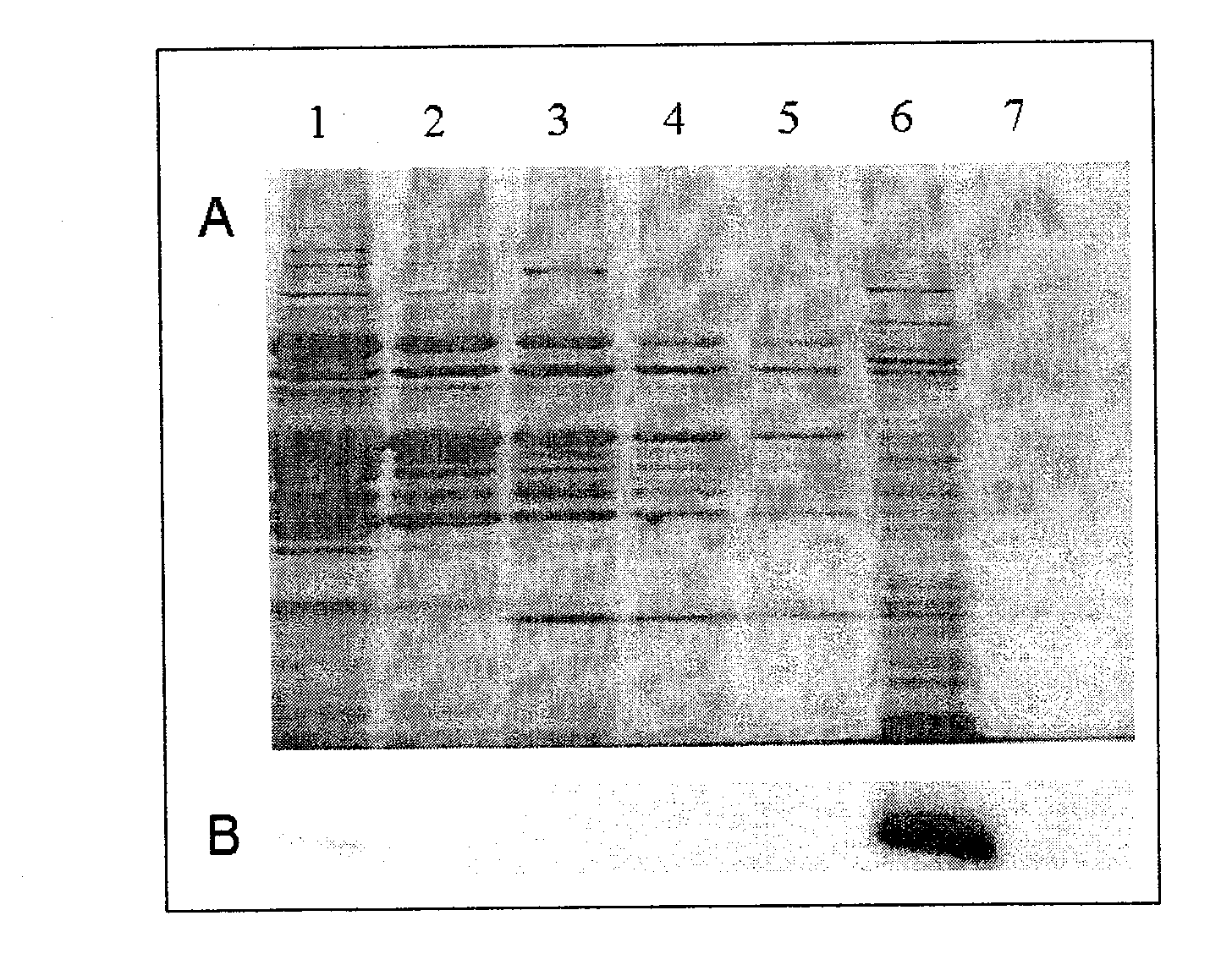
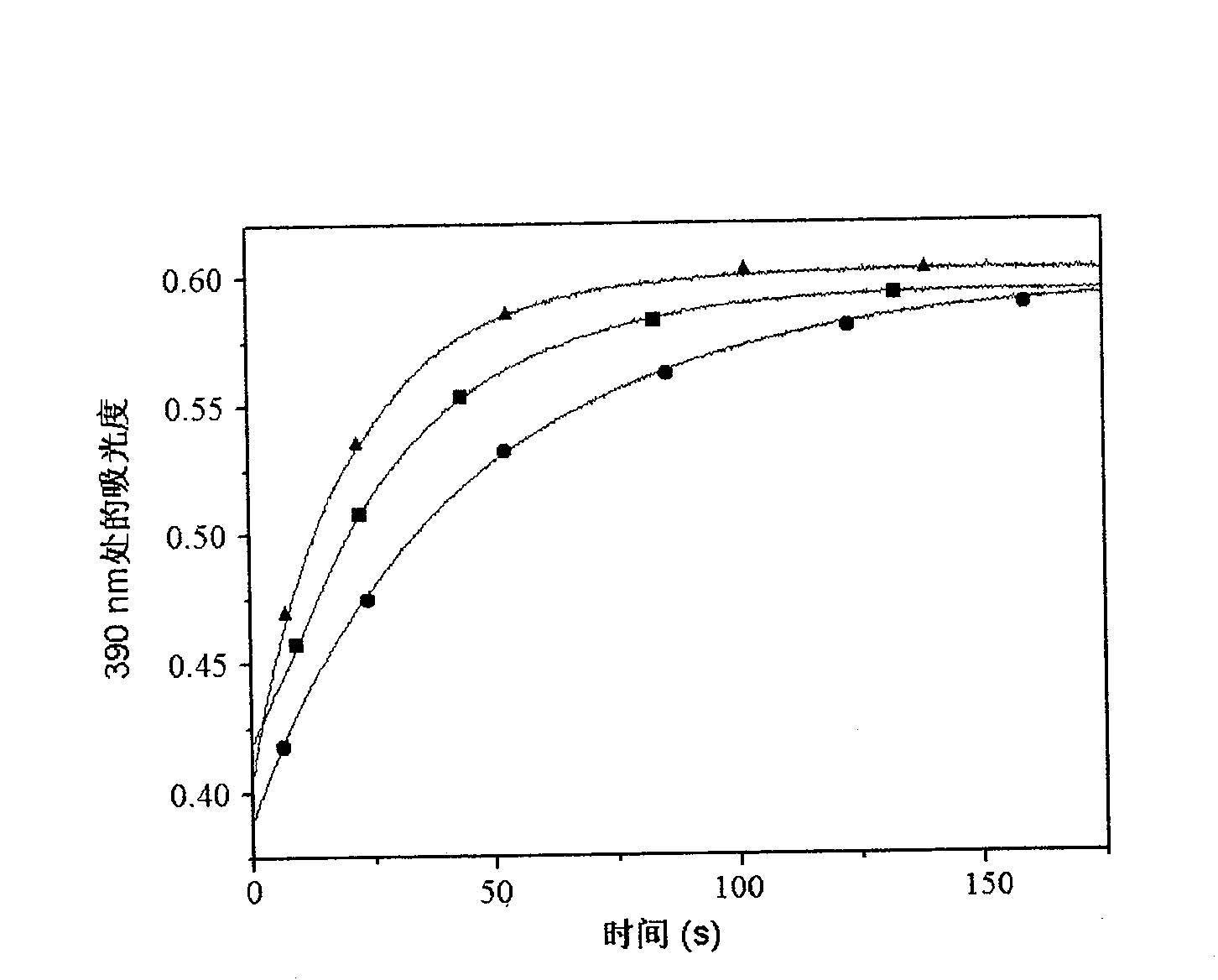
[0028] At 4°C, the above molecularly imprinted resin was mixed with 2 mL of porcine hepatocyte extract (cytosol), and the resin after 15 hours of spin adsorption was washed with 10 mM Tris-HCl (pH 8.0) buffer solution for 5 times, 2 mL each time, and then Wash twice with 2 mL buffer solution containing 10mM Tris-HCl (pH 8.0) and 100mM KCl, wash three times with buffer solution containing 10mM Tris-HCl (pH 8.0) and 300mM KCl, ) and a buffer solution of 500mM potassium chloride to wash the resin twice to obtain a washing solution. The Bradford method was used to detect the washing solution of potassium chloride solution and the pig liver cell extract solution to obtain the total protein content; then take 800 μL of the washing solution, and use the trichloroacetic acid / sodium deoxycholate (TCA / DOC) method to precipitate protein, protein After liquid sample preparation, constant flow sodium dodecylsulfon...
Embodiment 3
[0037] Embodiment 3, the preparation method of cloned protein porcine cyclophilin 18:
[0038] 1. Cloning of target gene
[0039] In this experiment, pig liver mRNA was used as a template for reverse transcription PCR (RT-PCR), and the PCR product and plasmid vector pGEX-5X1 were modified with restriction endonucleases. After ligation, they were transformed into E. coli DH5α and the plasmid was identified. .
[0040] (1) In a 200mL thin-walled PCR tube, add the required components in the following order for PCR reaction: 1 μL of RNase inhibitor, 100 ng of pig liver cell RNA, 25 μL of reaction buffer, and 10 pmol of forward primer. Reverse primer 10pmol, Taq mixed enzyme 1μL, double distilled water 21μL.
[0041] (2) Weigh 0.75g agarose, add 50mL 1×TAE, mix well and heat to boiling, take it out and add 2μL ethidium bromide (EB) solution (10mg / mL) when it cools slightly, pour the glue into Insert the comb into the gel stage of the electrophoresis tank; take out 16 μL of the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com