Establishment and application of recombined aminoglycoside dual-function modified enzyme in vitro metabolic model
A technology of aminoglycosides and modified enzymes, which is applied in the field of in vitro metabolic models of modified enzymes to antibiotics, can solve problems that have not been seen, and achieve the effect of simple method and high degree of automation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Cloning of embodiment 1 bifunctional modified enzyme gene aac (6')-aph (2 ")
[0041] (1) Extract total DNA from clinically isolated E.faecalis HH22:
[0042] Inoculate E.faecalis HH22 in Naoxintang medium containing 15 μg / mL gentamicin, shake and cultivate overnight at 200 rpm, centrifuge 2.5 mL of the bacterial liquid to enrich the bacterial cells, wash the precipitate twice with 500 μL STE, and then add 500 μL STE to refill To the suspension, add lysozyme to a final concentration of 2mg / mL, incubate at 37°C for 1h, then add 250μL of 2% SDS solution, shake and mix well, add proteinase K solution with a final concentration of 200μg / mL, incubate at 55°C for 30min, phenol / chloroform / isoamyl alcohol extraction until no denatured protein exists. Add 1 / 10 volume of 3M NaAc and an equal volume of isopropanol to the supernatant, and precipitate genomic DNA at room temperature.
[0043] (2) PCR amplification of the target gene:
[0044] The genomic DNA of E. faecalis HH22 ...
Embodiment 2
[0045] Cloning, sequencing analysis of the DNA fragment of embodiment 2 object
[0046] (1) Cloning the resulting bifunctional modified enzyme gene aac(6')-aph(2")TA into the pGEM-T vector
[0047] Purify the bifunctional modifying enzyme gene aac(6′)-aph(2″) obtained in Example 1 with a DNA purification kit, and use Taq enzyme to carry out an A-added reaction on the recovered PCR product, and then combine the A-added product with The pGEM-T vector was subjected to a ligation reaction to obtain a ligation product.
[0048] (2) Preparation of Competent Cells
[0049] Pick a single colony and inoculate in 2mL LB liquid medium, shake at 37°C and 200rpm overnight; inoculate 0.5mL overnight culture in 50mL LB liquid medium, shake at 37°C and 200rpm until OD 600 =0.3; the culture was ice-bathed for 10 min, centrifuged at 4°C and 4100 rpm for 10 min, and 0.6 times the volume of ice-precooled CaCl was used for precipitation 2 Wash once, then add 0.04 times the volume of ice-cold Ca...
Embodiment 3
[0052] Example 3 Construction of the bifunctional modified enzyme gene protein expression vector:
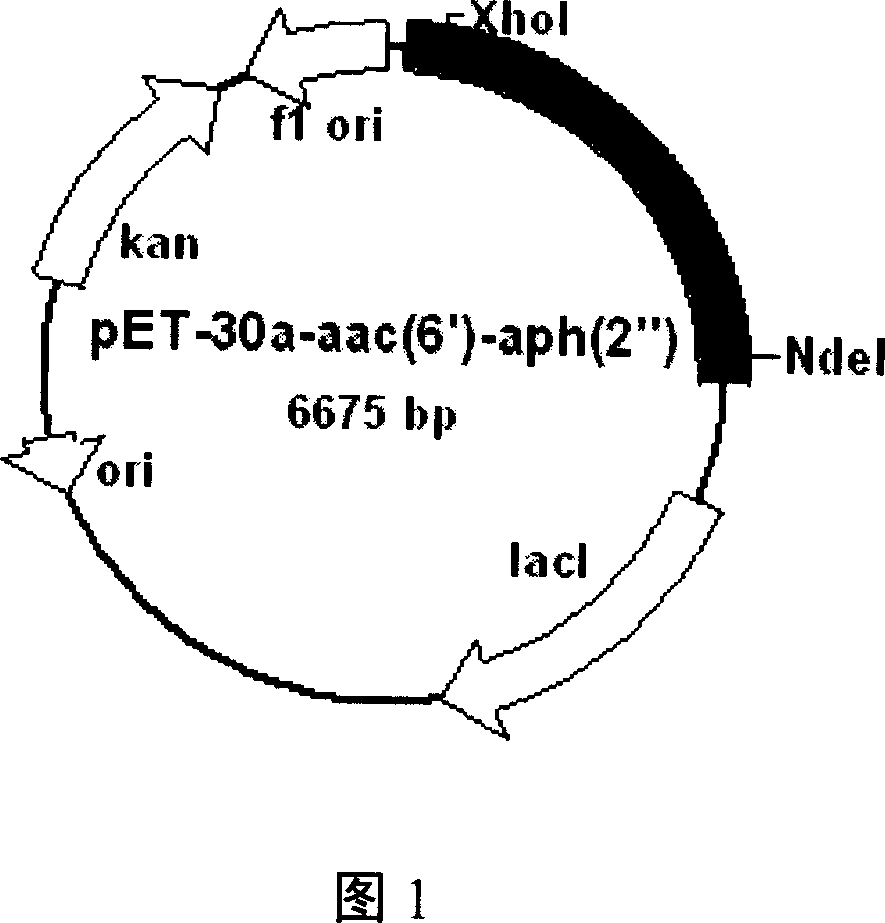
[0053] (1) The target fragment is connected into the expression vector
[0054] Using the enzyme cutting sites NdeI and XhoI introduced during the cloning of the target DNA fragment, cut it out from the pGEM-T vector, connect it to the protein expression vector pET-30a(+) after the same enzyme treatment, and transform it into the example The CaCl obtained by the method described in 2 2 The DH5α competent cells prepared by the method were coated with kanamycin-resistant plates, positive clones were selected, and the plasmids were extracted and identified by enzyme digestion, and sent to the sequencing company for sequencing.
[0055] (2) Transformation expression host
[0056] The Escherichia coli BL21 (DE3) competent cell prepared by the method described in Example 2 is transformed with the protein expression plasmid pET-30a-aac (6')-aph (2 ") that has been constructed; Coat k...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com