Method for detecting blood serum tumor markers
A tumor marker and detection method technology, applied in biological tests, measurement devices, material inspection products, etc., can solve the problem of lack of specific tumor markers for early screening of pancreatic cancer, and achieve improved cure rate, reasonable design, The effect of high specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
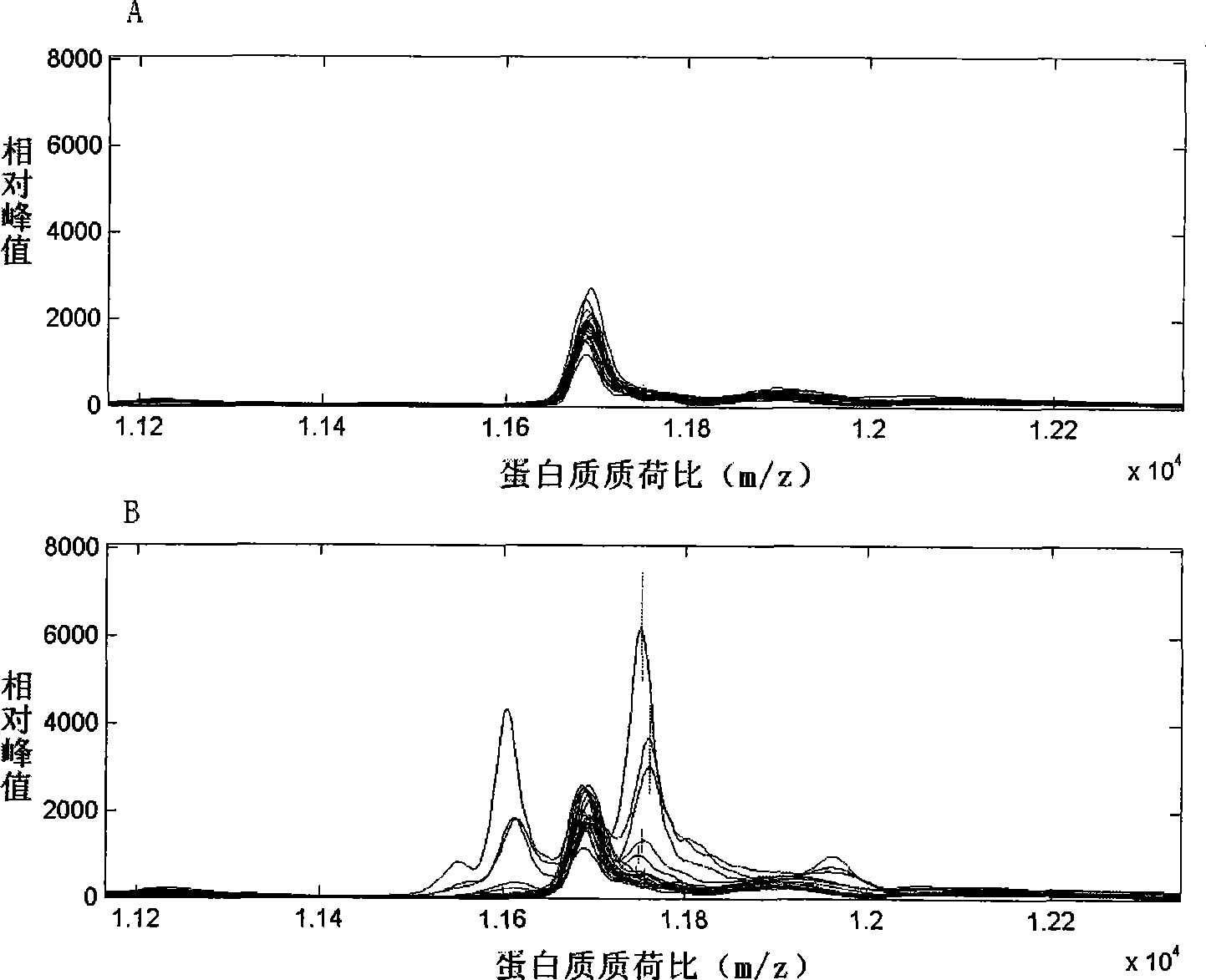
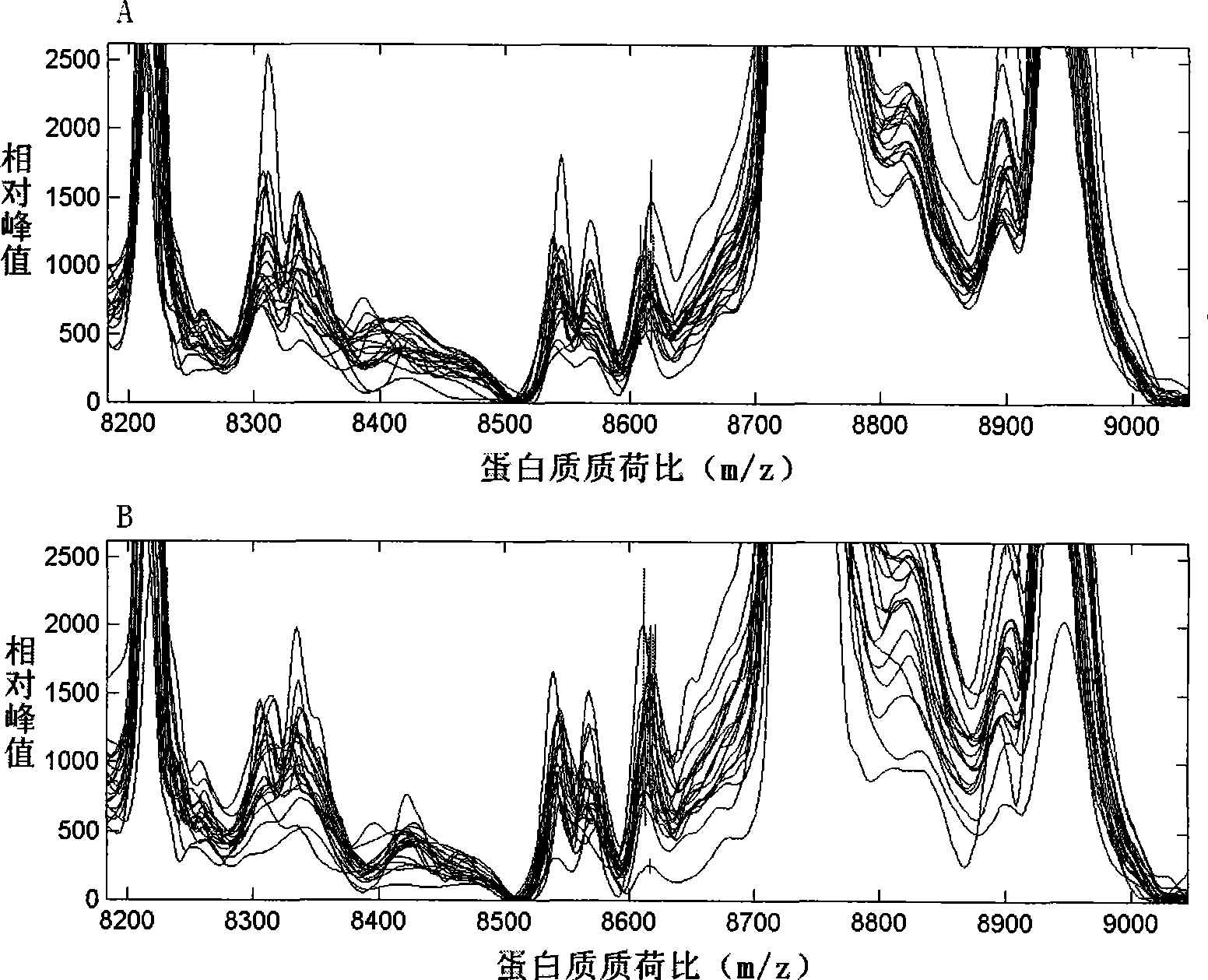
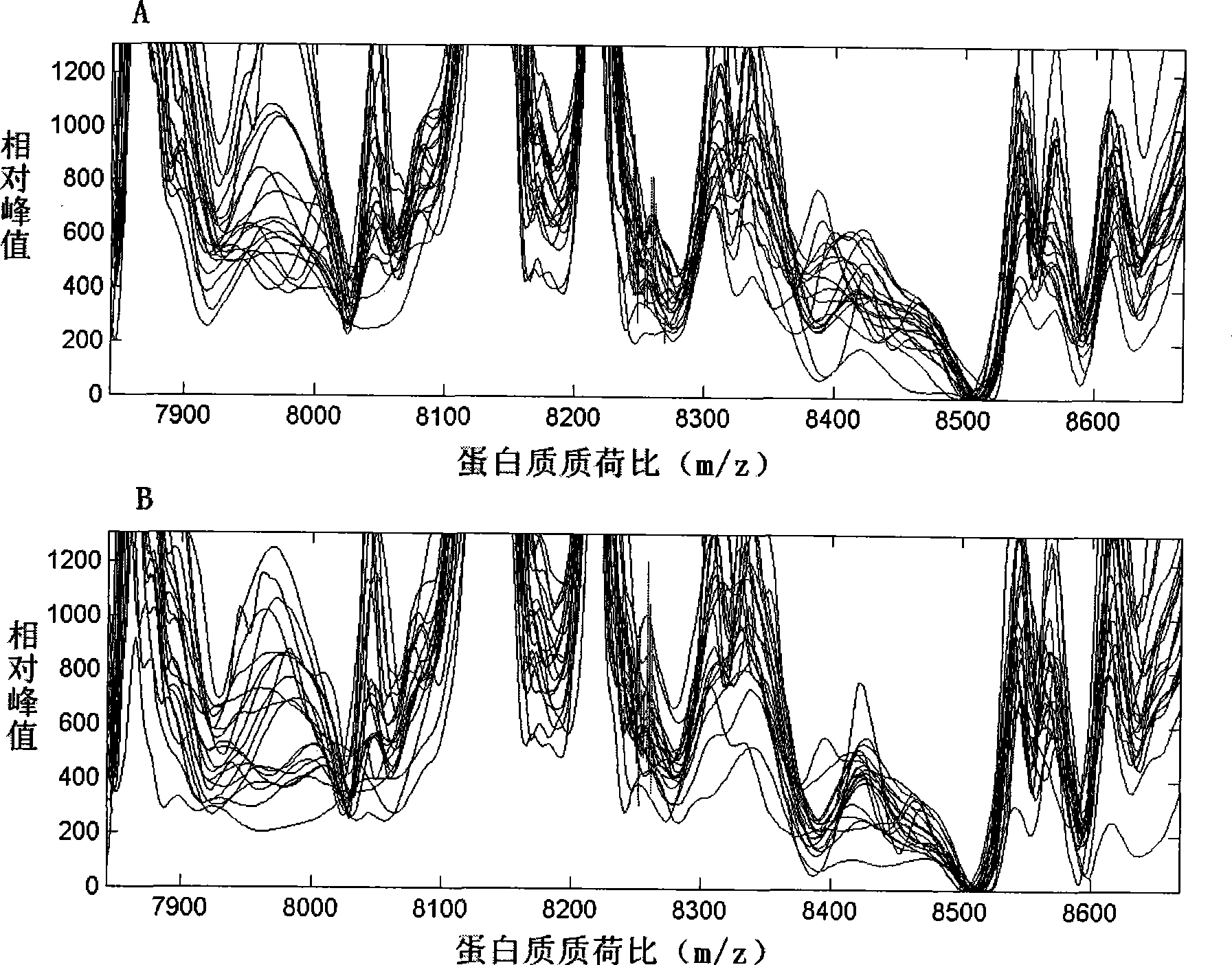
Image
Examples
Embodiment 1
[0019] Preparation of serum samples|:
[0020] Main instruments and reagents: CM10 protein chip
[0021] 96-well protein chip working support is Bio-proeessor
[0022] Analysis of pure urea, sodium acetate, acetonitrile, DTT and cHAPS buffer salt (sigma company)
[0023] Energy absorbing molecule SPA,
[0024] Implementation steps: take 1ml of whole blood, centrifuge at 4°C low temperature (4000 rpm) for 10 minutes, take the supernatant, centrifuge again for 10 minutes, and take the supernatant serum. Add 5 μL of serum to 10 μl of U9 serum treatment solution, shake on a shaker to mix thoroughly. U9-treated serum was diluted to 200 μl with Binding Buffer for loading.
[0025] Take 100 μl of the above-mentioned U9-treated and diluted serum and add it to the chip, and remove the sample after the chip fully reacts with the serum for 1 hour. Add 200 μl of corresponding Binding Buffer to each well, shake on a shaker for 5 minutes (600 rpm, room temperature), and then remove the...
Embodiment 2
[0027] Embodiment 2 On-machine detection:
[0028] Main instruments and software: PBS-II surface enhanced laser desorption ionization-time-of-flight mass spectrometer (SELDI-TOF-MS),
[0029]Analysis software ProteinChipSoftware3.0 (Ciphergen, USA): Support vector machines (supportveetor machines, SVM), discriminant analysis (discriminant analysis) and time-sequence analysis (time-sequence analysis) software are developed using Matlab6.5 data processing software (MathworkS company) ;
[0030] Implementation steps: mass spectrometer parameter setting: laser intensity 165, sensitivity 7, data collection range 2000-30000m / z (protein mass and charge ratio, ie mass-to-charge ratio), molecular weight is corrected with a standard protein chip before each data collection. Serum samples were detected by surface-enhanced laser desorption ionization-time-of-flight mass spectrometry to obtain serum protein profiles.
Embodiment 3
[0031] Example 3 Data Collection and Bioinformatics Analysis
[0032] The original data was first corrected with Proteinchip Software3.0 software to make the total ionic strength and molecular weight uniform, and to filter the noise. The initial noise filtering value was 5, and the secondary noise filtering value was 2. Clustering was performed with 10% as the minimum threshold value. After the above data preprocessing, the Mann-Whitney rank test was used to compare the serum protein mass spectrometry data of each group (completed by the Biomarker Wizard3.2.0 software that comes with Proteinchip Software3.0), to find protein peaks with differences in expression among the groups.
[0033] The data of serum protein mass spectrometry comparison in each group were imported into the SVM and discriminant analysis software of Matlab6.5 data processing software. The 10 protein peak data with the most significant difference between the two groups were selected for analysis. The combin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com