Biochip for detecting drug resistant genes of helicobacter pylori clarithromycin and preparation method and application thereof
A technology of Helicobacter pylori clarithromycin and biochip, which is applied in the fields of genetic engineering and medical testing, can solve problems such as low detection cost, achieve the effects of high sensitivity, reduced detection cost and simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1, preparation biochip
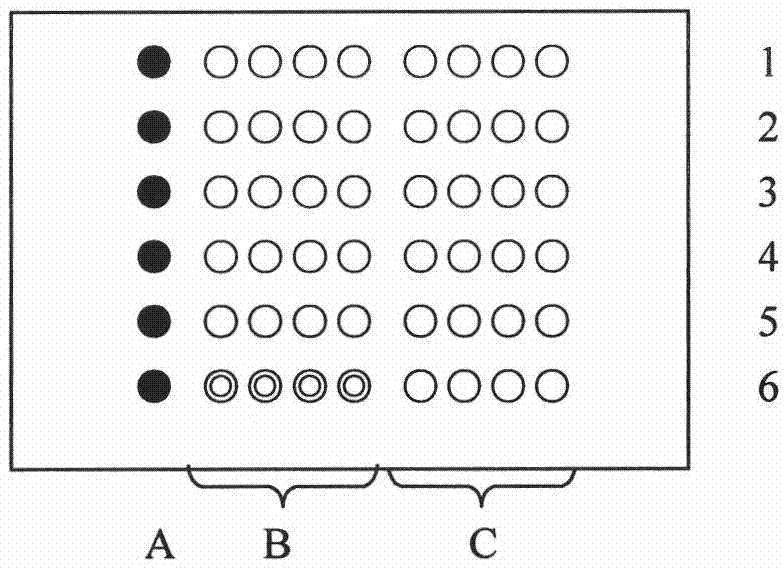
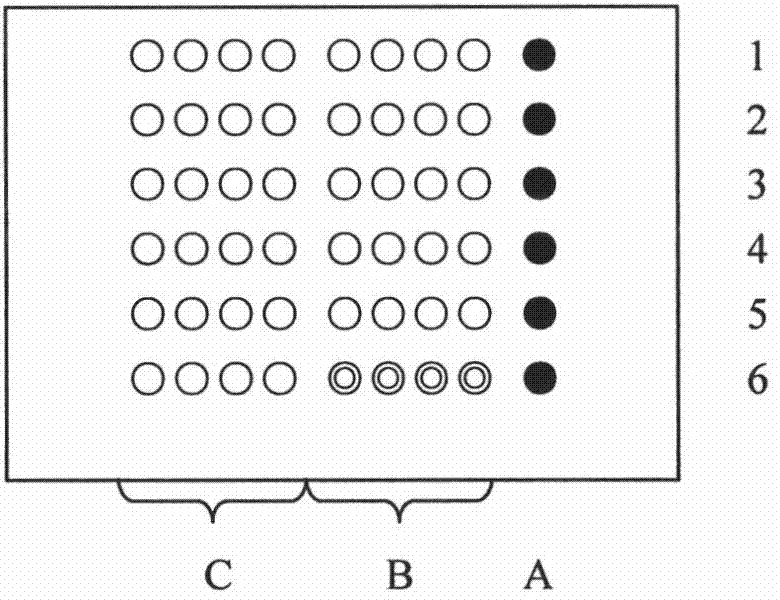
[0055] The chip base (glass slide) is treated with aldehydes to facilitate the fixation of the probe; the probe sequence designed according to the point mutation (2115, 2142, 2143, 2182, 2224) of the Helicobacter pylori 23S rRNA gene, the probe The 5' end is connected to poly(dT) 10 to increase the distance between the probe and the chip base, and reduce the steric hindrance during hybridization, and the 5' end of the probe is then modified with an amino group (see Table 1) so as to be compatible with the aldehyde group on the chip base. Generate coupling to make the probe easier to immobilize on the chip base;
[0056] Table 1 The probe sequence designed according to the point mutation of Helicobacter pylori 23S rRNA gene
[0057]
[0058] Dilute each synthesized probe to 300 pmol / μl with deionized water, take 10 μl spotting solution (750 mmol / L NaCl, 75 mmol / L sodium acetate, 5% glycerol, 1% polysucrose and 0.1% SDS) and 10 μl ...
Embodiment 2
[0059] Example 2, Digoxigenin labeling of Helicobacter pylori DNA fragments
[0060] Gastric mucosal DNA was extracted using TaKaRa’s DNA extraction kit (DV811A, operated strictly according to the instructions), and the extracted DNA was stored at -30°C as a template for future use. A pair of primers were designed according to the 23S rRNA mutation site (primer synthesis was completed by Shanghai Sangon Biotechnology Co., Ltd.), the primer sequences were: 5′-ATGAATGGCGTAACGAG-ATGGGAGC-3′ and 5′-CCAGTCAAACTACCCACCAAGCATT-3′, and the upstream primers were labeled with digoxigenin . PCR amplification produces DIG-labeled target DNA fragments. 50μl PCR amplification reaction system contains 1.5mmol MgCl 2 , 200 μmol of dNTP, 5 μl of template DNA, 1x buffer, 1 U Taq enzyme, 0.1 μmol of upstream and downstream primer concentrations. PCR amplification conditions: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 sec, annealing at 53°C for 30 sec, extension at 72°C fo...
Embodiment 3
[0061] Embodiment 3, the hybridization of labeled target DNA fragment and chip
[0062] Take 1 μl of the above-mentioned PCR-amplified Digoxigenin-labeled target DNA sample and 10 μl of hybridization solution (750mmol / L NaCl, 75mmol / L sodium acetate, 0.1% SDS, 1ug / ml salmon sperm DNA, 25% formamide) and mix at 94°C After denaturation for 5 minutes, quickly cool on ice for 5 minutes, take 10 μl of the mixture and add it dropwise to the spotting area of the chip, cover with a cover glass, place the chip in a 37°C preheated hybridization box, incubate in an oven for 30 minutes, and then preheat it at 60°C Washing solution I (0.2% SDS, 150mmol / LNaCl, 150mmol / L sodium acetate) shaking and washing (60-100 times / min) for 10min, washing solution II (150mmol / L maleic acid, 150mmol / LNaCl, 0.3% Tween20, pH 7.5) rinse for 1min and dry, add 2500 times diluted enzyme-labeled antibody (alkaline phosphatase-labeled digoxin antibody, Roche product) 20μl in the spotting area, let stand at 37°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com