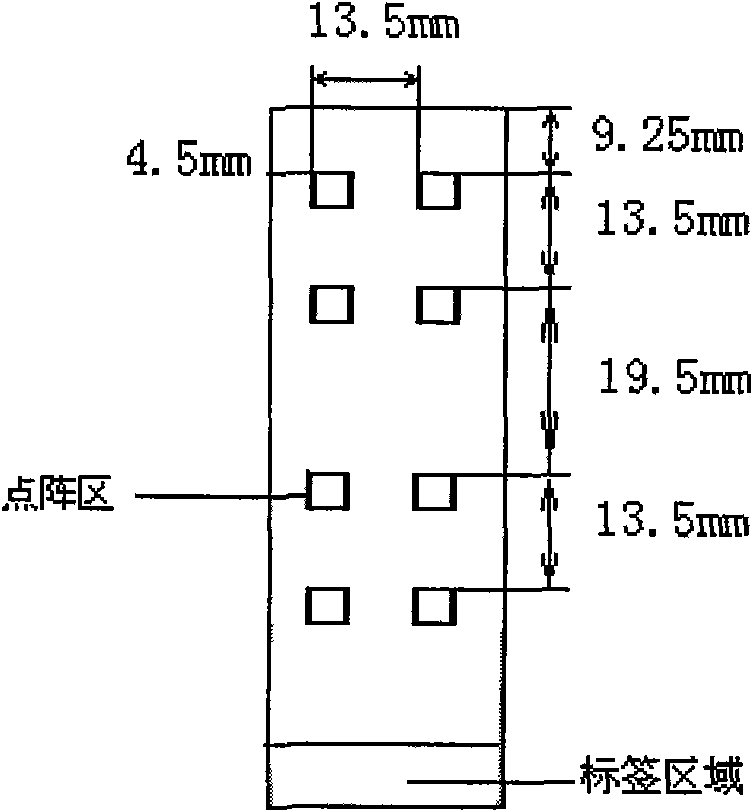
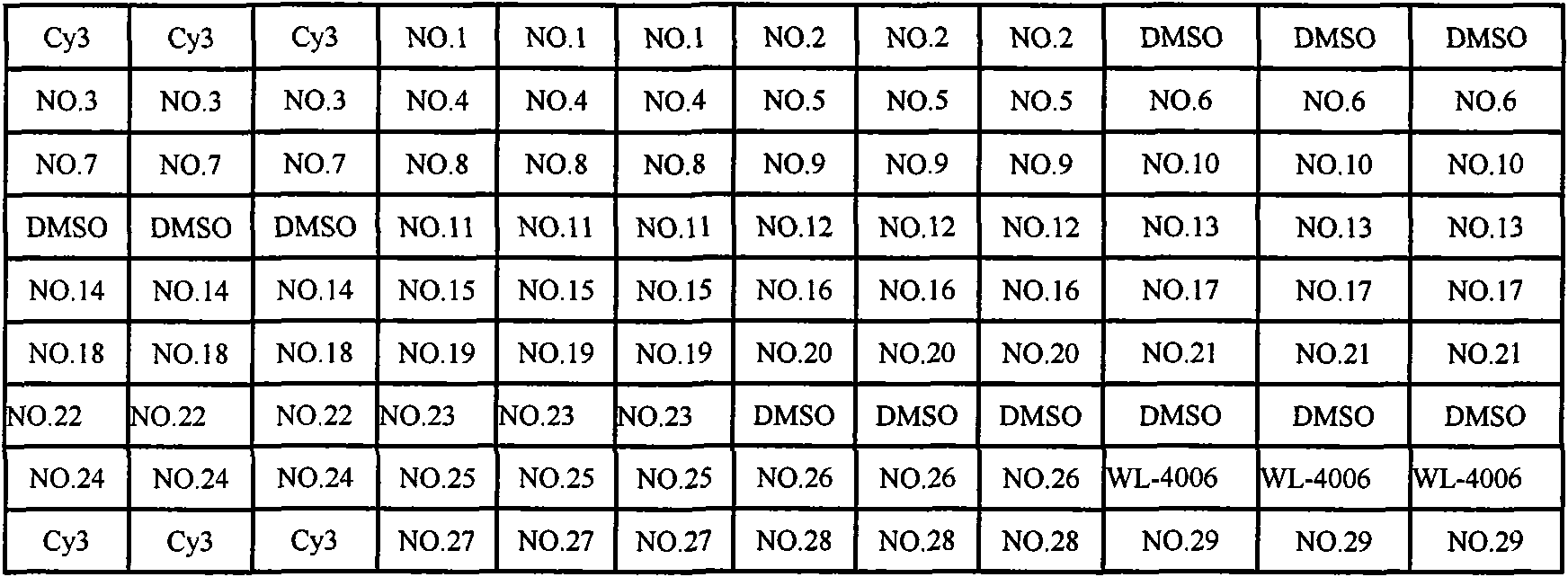
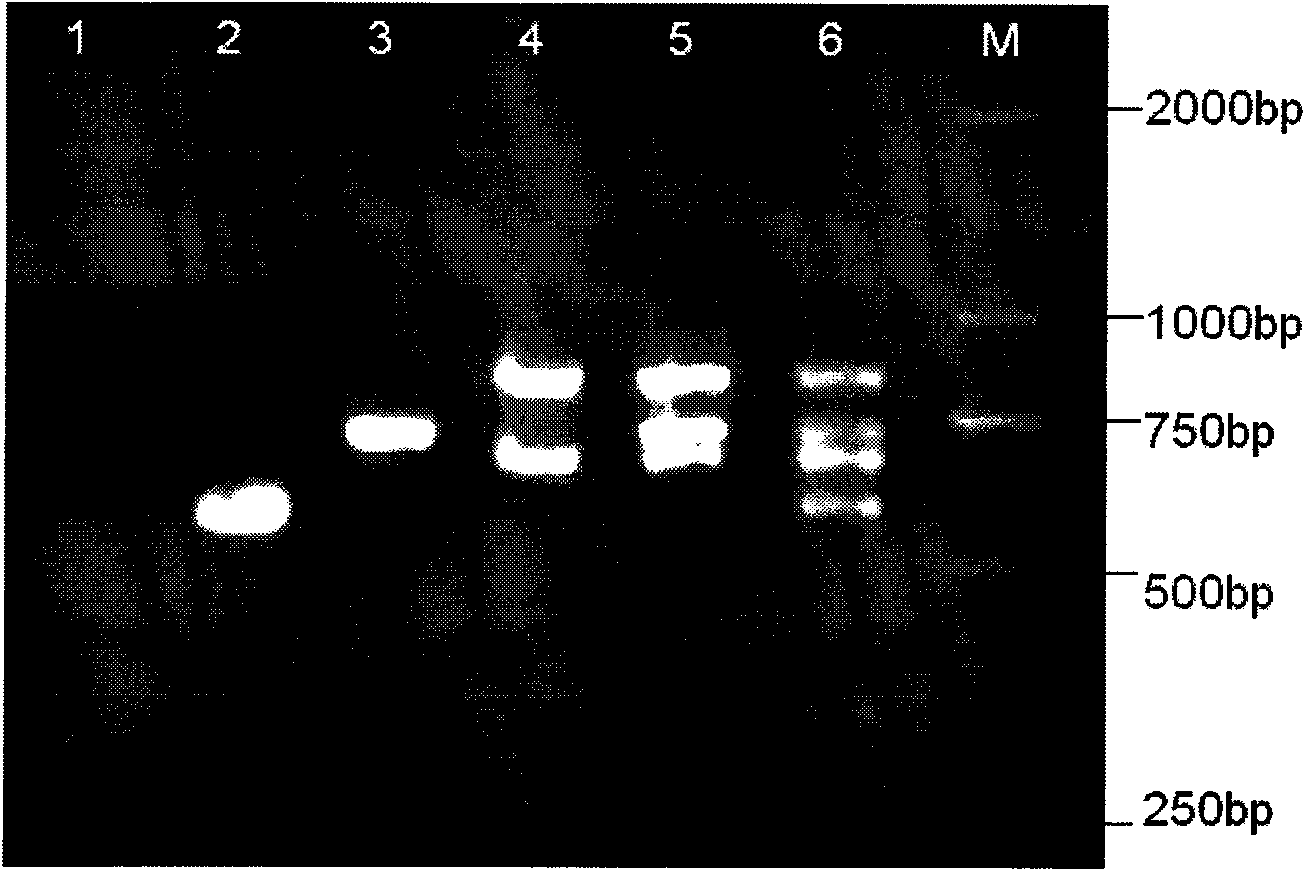
Gene chip for detecting important drug resistant gene of pseudomonas aeruginosa and kit thereof
A technology of Pseudomonas aeruginosa and drug-resistant genes, which is applied in the direction of microbial-based methods, microbial measurement/testing, chemical libraries, etc., can solve the problem of high chip manufacturing cost, achieve low cost, cost reduction, and good stability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Probe Design and Preparation
[0050] 1. Sequence to get:
[0051] The entire gene sequence of the above-mentioned drug resistance genes of Pseudomonas aeruginosa and the typical sequence of bacterial 16S rRNA gene were downloaded from the GenBank public database.
[0052] 2. Probe design:
[0053] Clustal X software was used to compare the different sequences of each gene. According to the comparison results, a representative sequence was selected and imported into Primer Premier 5 software. Run the program to design the probe, select the length (25 ± 5) bp, T m 60 ℃ ~ 70 ℃, GC content 40% ~ 60%, the designed probe was imported into NCBI for blastn comparison to ensure the intragene conservation and intergene specificity of the sequence. At least 4 different probes were designed for each gene for subsequent screening.
[0054] 3. Probe synthesis: The designed probe sequence is added to the total length of 40 nucleotides (positive control probe is lengt...
Embodiment 2
[0061] Embodiment 2 Primer design and preparation
[0062] 1. Examples of PCR primer design:
[0063] (1) Example of the design of universal amplification primers for a single drug resistance gene TEM: Import all TEM sequences of Pseudomonas aeruginosa (including each TEM genotype) into Glustal X software, and select a representative sequence from it and import it into Primer Primer 5.0 In the software, the length is set to (20±4) bp, the G+C% value is 40% to 60%, and the product size is greater than 200 bp. And find the nucleotide sequence region suitable for the design of universal primers, and its characteristics must meet the following conditions: 1, the constant region should include most TEM types; 2, the region should contain a region that is easy to design specific probes 3. The constant regions on both sides of this region can meet the design of primers. The primer sequence with the smallest secondary structure (hairpin structure, dimer) was selected, and the sequ...
Embodiment 3
[0069] Embodiment 3 Detection of 10 common important drug resistance genes of Pseudomonas aeruginosa by gene chip and preparation of the kit
[0070] 1. Pre-hybridization of the chip
[0071] (1) Put the ordered chip into 500 ml of pre-hybridization solution preheated at 42°C, and incubate at 42°C for 1 hour.
[0072] Pre-hybridization solution formula: 1% BSA (bovine serum albumin)
[0073] 5×SSC (sodium chloride-sodium citrate solution)
[0074] 0.1% SDS (sodium dodecyl sulfate)
[0075] (2) at ddH 2 Washed in O medium twice for 1 minute, dehydrated in 95% ethanol for 2 minutes, dried with a hair dryer, and set aside.
[0076] 2. Obtaining total DNA:
[0077] (1) The collected Pseudomonas aeruginosa was picked up and the monoclonal colony was transferred into 5ml of 2YT culture solution for 37°C shaking culture for 12 hours;
[0078] (2) Take 1ml of bacterial liquid into a 1.5ml centrifuge tube, centrifuge at 15000g × room temperature for 3 minutes;
[0079] (3) Dis...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com