Method for producing L-ornithine hydrochloride by genetic engineering bacteria
A technology of ornithine hydrochloride and genetically engineered bacteria, which is applied in the biological field and can solve problems such as filtration difficulties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
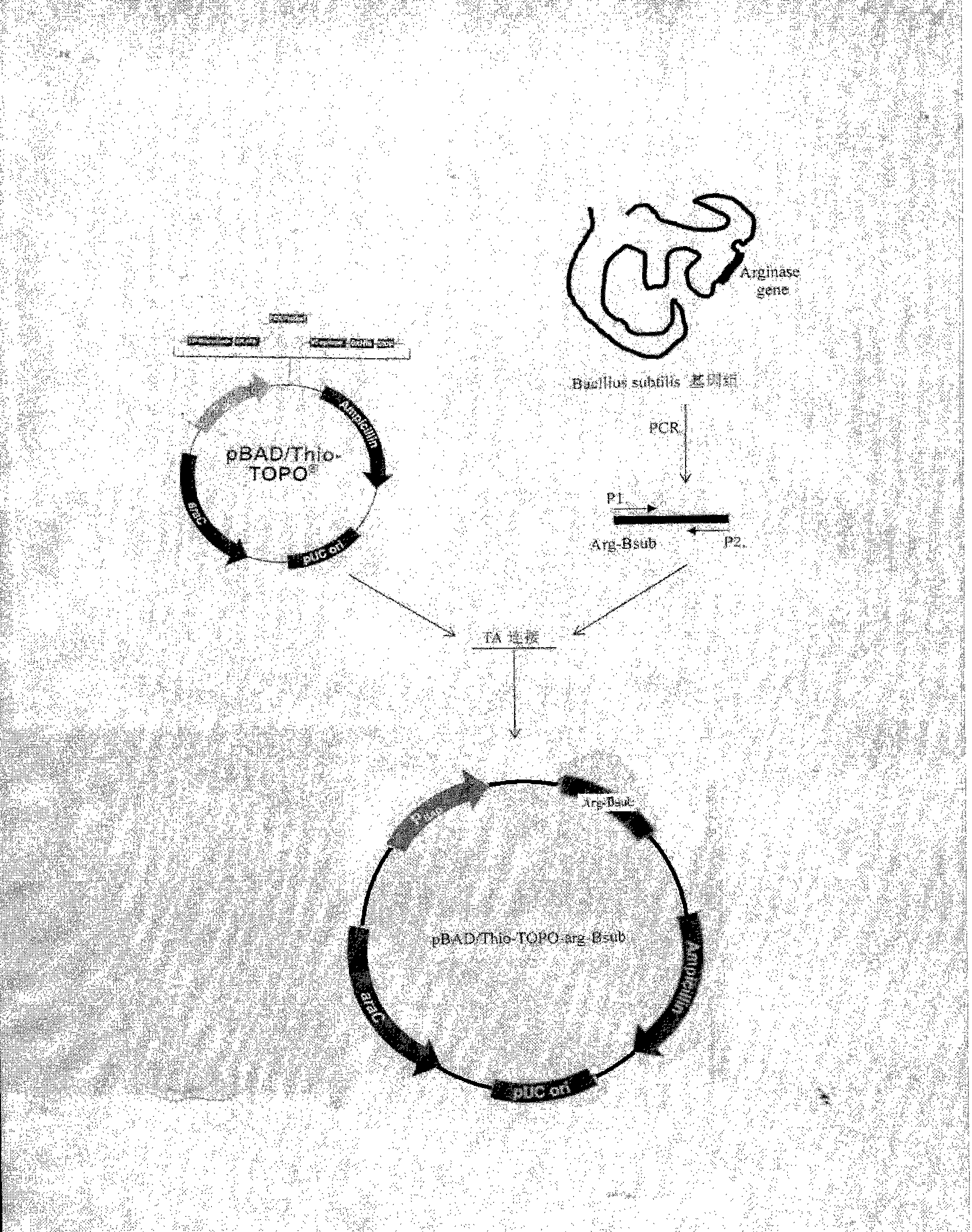
[0180] Design and synthesize a pair of TA cloning primers
[0181] Log in to GenBanK, obtain the complete coding sequence of the arginase gene (argBsub) of Bacillus subtilis (Bacillus Subtilis) 168 strain through Gene ID: 937760, and use Primerer 5.0 software to design a pair of PCR primers, p1 and p2, for amplifying subtilis The DNA sequence of the Bacillus arginase gene is TA-ligated with the pBAD / Thio-TOPO vector. Primers were synthesized by Yingwei Jieji (Shanghai) Trading Co., Ltd.
[0182] P1: 5'-GAAATGGATAAAACGATTTCGGTTAT-3', 26 bp in total.
[0183] P2: 5'-GGTCAGCAGCTTCTTCCCTAACA-3', 23 bp in total.
Embodiment 2
[0185] Extraction and determination of Bacillus subtilis genomic DNA
[0186] Genomic DNA of Bacillus subtilis was extracted by the improved method of Palva I et al.
[0187] (1). Pick a single bacterium colony cultured on the plate of Bacillus subtilis (preservation number CCTCC NO.AB93009), inoculate it into a test tube containing 10 ml of beef extract and peptone liquid medium, and culture it with shaking at 32° C. for 14 hours.
[0188] (2). Centrifuge at 5000rpm for 10min to obtain bacterial pellet, wash once with STE, centrifuge again, and resuspend the bacterial cell in 4ml TE solution.
[0189] (3). Add 8 μl of 50 mg / ml lysozyme solution (final concentration is 100 μg / ml), and keep warm at 37° C. for 20 minutes. Add 10 μl of RNase (10 mg / ml) to a final concentration of 25 μg / ml, then add 10% SDS to dissolve 0.5 ml, and incubate at 37° C. for 30 minutes. Add 10 μl of proteinase K (20 mg / ml) to a final concentration of 50 μg / ml, and keep at 37° C. for 60 minutes.
[0...
Embodiment 3
[0199] PCR Amplification of Arginase Gene of Bacillus subtilis
[0200] 1). Prepare 50 μl reaction system.
[0201] Template DNA, 2μl (50ng); 10X PCR Buf, 5μl; 50mMdNTPs, 0.5μl;
[0202] Primers p1 and p2, 1 μl each (100 ng); Taq DNA polymerase (1 unit / μl), 1 μl; add sterile pure water to a volume of 50 μl.
[0203] The program of the PCR reaction was as follows: pre-denaturation at 94°C for 4 min, denaturation at 94°C for 1 min, annealing at 56°C for 30 sec, extension at 72°C for 1.5 min; a total of 30 cycles, and a final extension at 72°C for 10 min. After the reaction was completed, temporarily store in ice until use.
[0204] 2). Take 2 μl of the amplification product and load it on 1.2% agarose gel electrophoresis to detect a DNA fragment with a size of 0.9 KB. The DNA band has a clear outline, which is suitable for direct TOPO-TA cloning reaction.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com