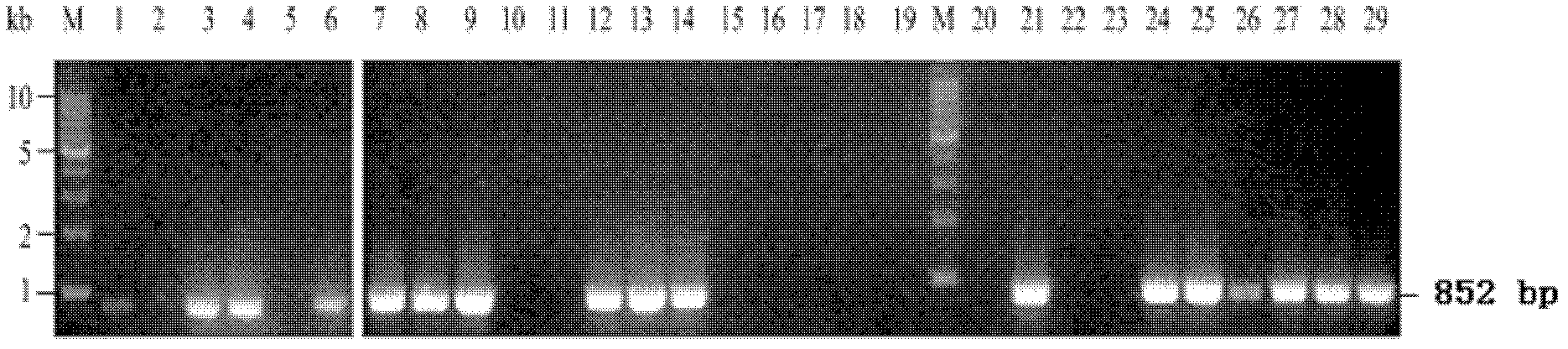Method and kit for screening/detecting lactobacillus
A Lactobacillus and kit technology, applied in the field of bacterial detection, can solve the problems of high cost, huge workload, strong primer specificity, etc., and achieve the effect of strong application value, cost saving and time saving.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Primer Design
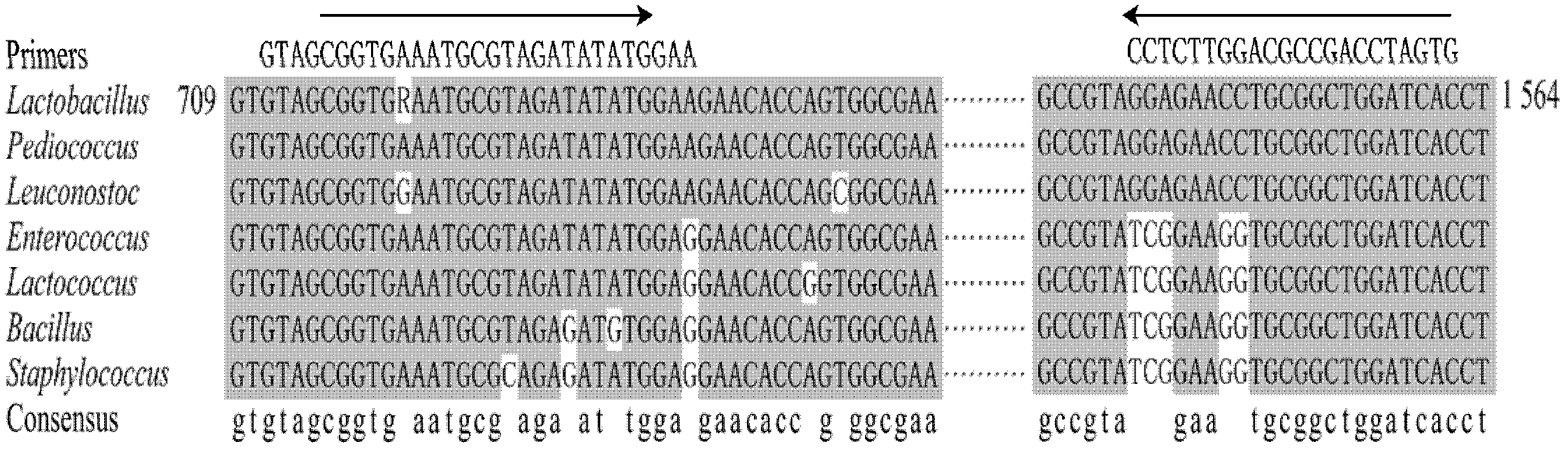
[0025] As shown in Table 1, according to 14 kinds of Lactobacillus genome sequences in the Genome database of NCBI, look for the Lactobacillus conserved sequence, this conserved sequence and Lactobacillus belong to the same Lactobacillus Pediococcus (Pediococcus), and Lactobacillus belong to the same Lactobacillus order Enterococcus (Enterococcus), Lactococcus (Lactococcus) and Leuconostoc (Leuconostoc), and Bacillus (Bacillus) and Staphylococcus (Staphylococcus), which belong to the same genus of Lactobacillus, are aligned with the corresponding sequences, and Lactobacillus-specific primers are designed .
[0026] Table 1 List of 14 Lactobacillus species and 5 closely related species that have been sequenced
[0027]
[0028] In order to achieve the purpose of quickly screening out all lactobacilli from a sample containing a large number of microorganisms, a pair of primers needs to be designed in the present invention, and the primers sho...
Embodiment 2
[0032] Example 2 Primer Specificity Experiment
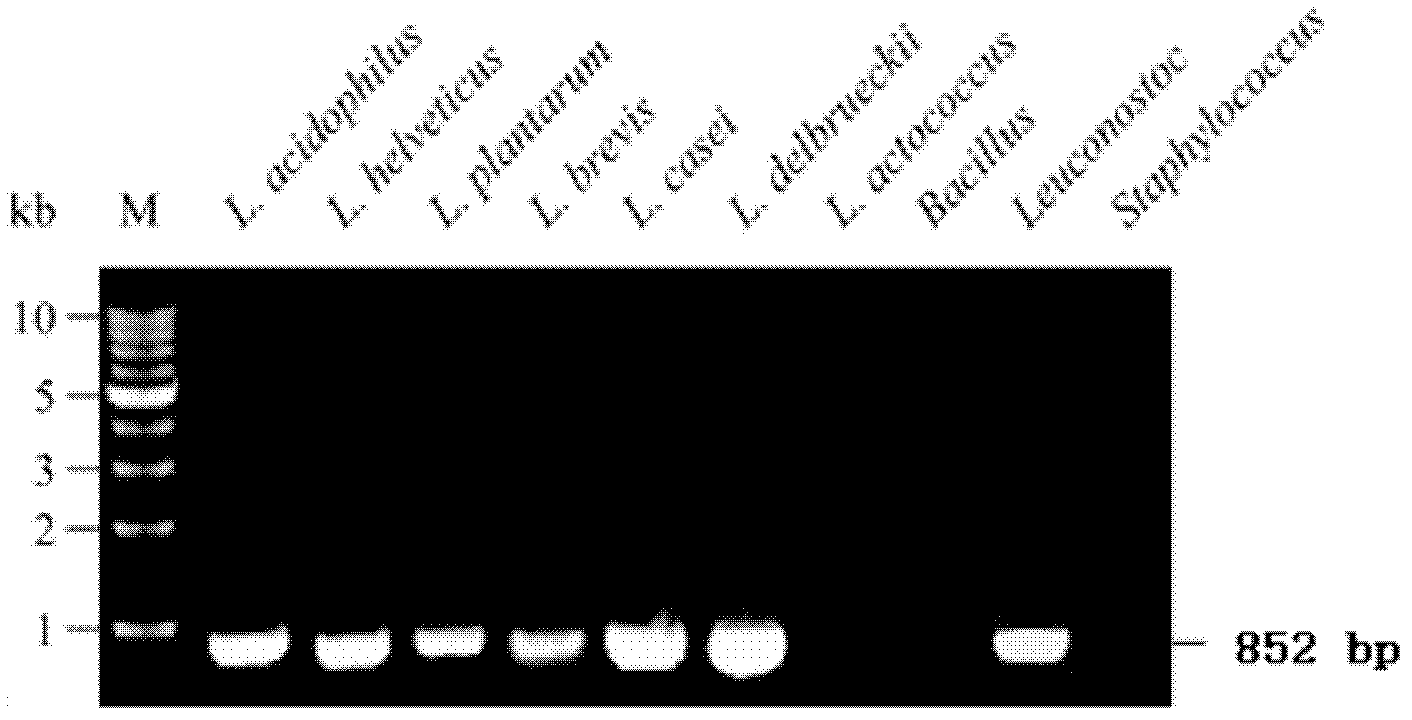
[0033] For Lactobacillus acidophilus, Lactobacillus plantarum, Lactobacillus brevis, Lactobacillus casei, Lactobacillus helveticus, Lactobacillus delbrueckii, Leuconostoc, Lactococcus lactis, Bacillus subtilis and Staphylococcus epidermidis, PCR amplification detection was carried out as follows :
[0034] 1. Primers
[0035] The primers prepared in Example 1 were used.
[0036] 2. Template preparation
[0037] Pick traces from single colonies of Lactobacillus acidophilus, Lactobacillus plantarum, Lactobacillus brevis, Lactobacillus casei, Lactobacillus helveticus, Lactobacillus delbrueckii, Leuconostoc, Lactococcus lactis, Bacillus subtilis and Staphylococcus epidermidis, respectively The bacterial cells were placed in a PCR tube, the PCR tube was placed in a microwave oven, heated at 100% fire for 3 min, and then added to the PCR system.
[0038] 3. PCR amplification
[0039] Using the heat-treated cells in step 2 as temp...
Embodiment 3
[0053] Embodiment 3 The composition of the kit of the present invention and its use method
[0054] 1. Composition of the kit
[0055] 25μL system:
[0056] Primers provided in Example 1: 0.5 μl each
[0057] 10x PCR buffer: 2.5 μl
[0058] Magnesium Chloride: 2 µl
[0059] dNTP: 2 µl
[0060] exTaq enzyme: 0.25 μl
[0061] ddH 2 O: 17.25 µl
[0062] 2. How to use the kit
[0063] 1), isolate a single colony: isolate a single colony of strains from the sample to be tested;
[0064] 2), gene amplification:
[0065] Using the bacterial genome DNA obtained in step 1) as a template, gene amplification was performed with a kit. The reaction procedure was as follows: 94°C for 5 min; 95°C for 40s, 57°C for 30s, 72°C for 100s, 30 cycles; 72°C for 10 min.
[0066] 3), result detection:
[0067] 5 μL of the reaction solution was taken and subjected to agarose gel (0.7% W / V) electrophoresis to detect PCR amplification results.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com