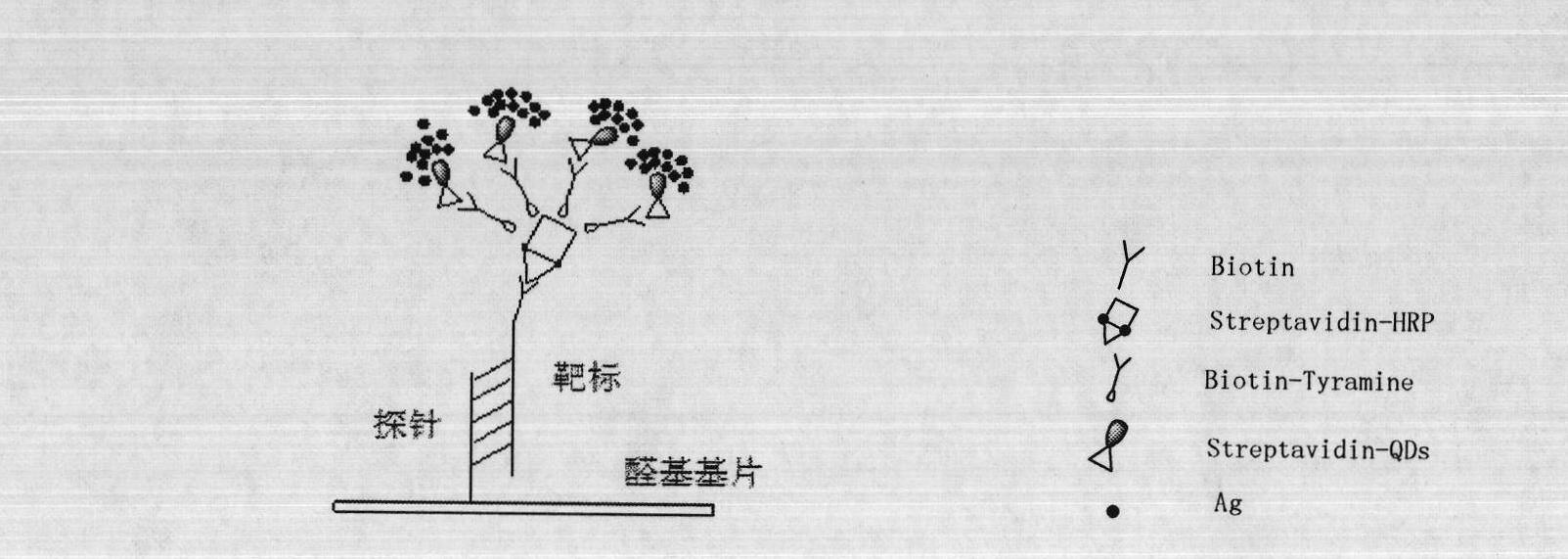
Molecule detection signal amplification technique
A technology for detecting signals and signal amplification, applied in biological testing, material inspection products, etc., can solve the problems of expensive detection instruments, easy saturation of detection signals, poor signal stability, etc., to achieve visual detection, stable detection signals, and high detection. Effects of sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
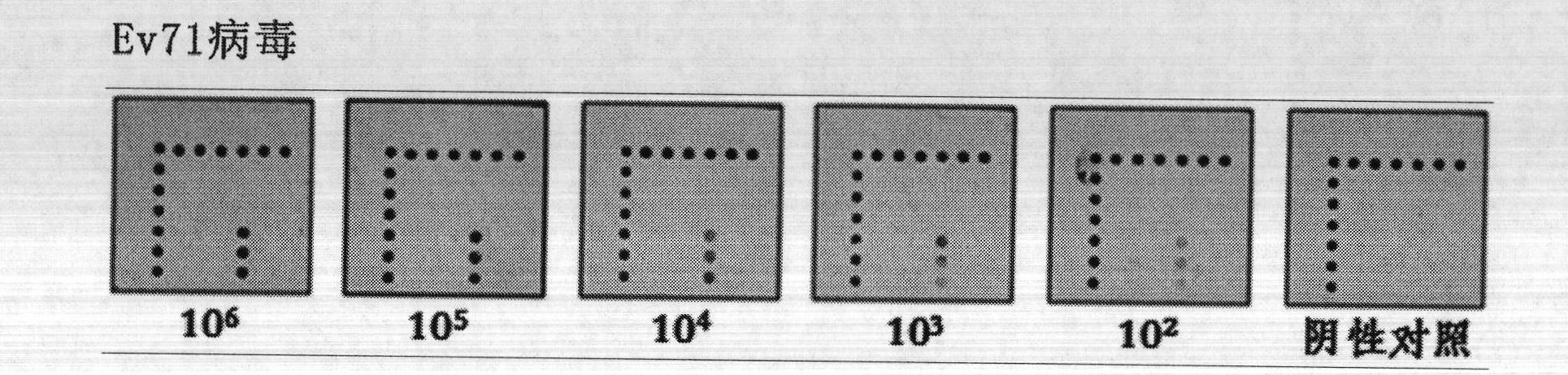
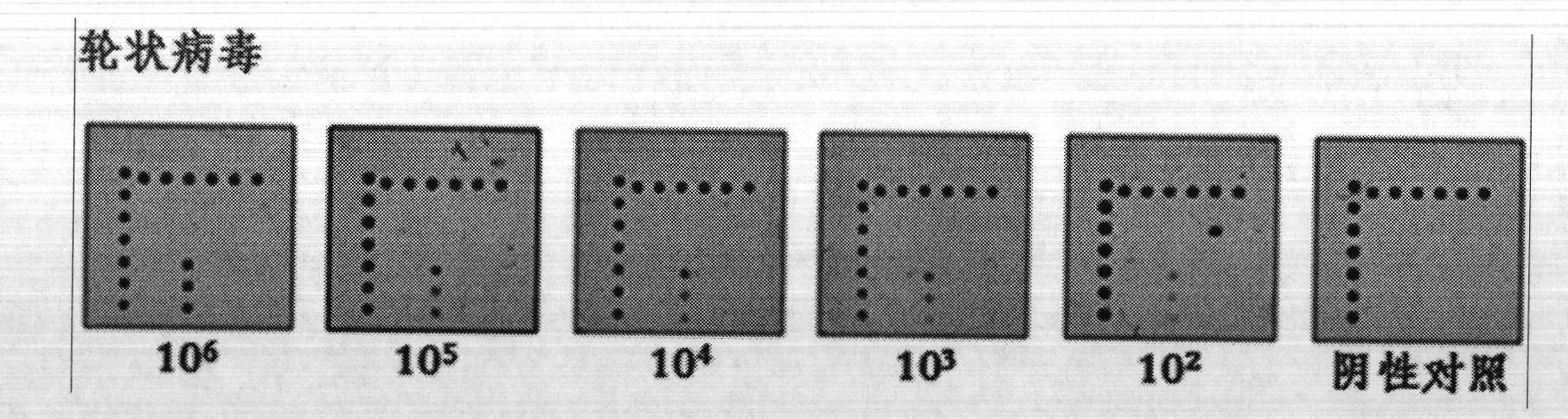
[0030] Detection of Ev71 virus and rotavirus
[0031] 1. Self-made gastrointestinal virus detection gene chip.
[0032] 2. The Ev71 virus and rotavirus cell culture medium were operated according to the instructions of the QIAamp Viral RNA Mini Kit Nucleic Acid Extraction Kit (QIAGEN) to obtain purified RNA. Use the reverse transcription kit PrimeScript One Step RT-PCR Kit Ver.2 (Bao Biological Engineering Co., Ltd.) for reverse transcription, the reaction system is 20 μL: 2 × One Step Buffer 10 μL, PrimeScript 1 Step Enzyme Mix 0.8 μL, 0.2 μL ( Upstream primer, 10 μM), 1 μL (downstream primer, 10 μM), RNA template 5 μL, nuclease-free water to make up the volume to 20 μL system. Reaction conditions: reverse transcription: 50°C, 30min; pre-denaturation: 94°C, 2min; PCR amplification: 94°C, 30s; 50°C, 30s; 72°C, 30s; extension: 72°C, 10min. A total of 45 cycles were performed in PCR amplification.
[0033] 3. Wash the prepared gene chip with 0.2% SDS for 20 seconds, then with...
Embodiment 2
[0039] Detection of Coxsackie B3, Coxsackie B4, Coxsackie B5, Polio1, Polio2, Polio3, Eike 30, Coxsackie A16, Enterovirus Virus type 71, rotavirus.
[0040] Self-made gene chip for gastrointestinal virus detection.
[0041] 1. Coxsackie B3 virus, Cossackie B4 virus, Cossackie B5 virus, polio1 virus, polio2 virus, polio3 virus, Eike 30 virus, Cossackie A16 virus, Enterovirus 71 and rotavirus cell culture medium were operated according to the instructions of the QIAamp Viral RNA Mini Kit Nucleic Acid Extraction Kit (QIAGEN) to obtain a purified RNA solution. Use the reverse transcription kit PrimeScript One Step RT-PCR Kit Ver.2 (Bao Biological Engineering Co., Ltd.) for reverse transcription, the reaction system is 20 μL: 10 μL (2×One Step Buffer), 0.8 μL (PrimeScript 1Step Enzyme Mix) , 0.2 μL (upstream primer, 10 μM), 1 μL (downstream primer, 10 μM), 5 μL (LRNA template), and nuclease-free water to make up the volume to 20 μL. Reaction conditions: reverse transcription: 50...
Embodiment 3
[0048] Detection of hepatitis B surface antigen HBsAg in patients' serum
[0049] Chip preparation: HBsAb (0.5 mg / mL) was spotted on the aldehyde-modified glass slide; BSA (1 mg / mL) negative control. After being placed at 4°C for 12h, it was blocked with blocking solution (1×PBS+25% calf serum) for 2h, then washed three times with PBST (1×PBS+0.5% Tween) and dried at 4°C for later use.
[0050] 1. Take the prepared chip and add sample serum (1:5 dilution) in sequence, incubate at 37°C for 30 minutes, take out the chip and wash it three times with PBST (1×PBS+0.05% Tween20) for 20 seconds each time, and dry it in the air.
[0051] 2. Add HbsAb-Biotin (1:1000, 1×PBS-0.5%BSA, 1mg / mL) to the corresponding area and incubate at 37°C for 30min, take out the chip and wash it three times with PBST (1×PBS+0.05%Tween20) repeatedly 20s, dry.
[0052] 3. Add Streptavidin-QDs (diluted 1:50, 2XPBS-0.1% BSA-0.01% thimerosal) in the corresponding area, incubate at 37°C for 30min, wash with P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com