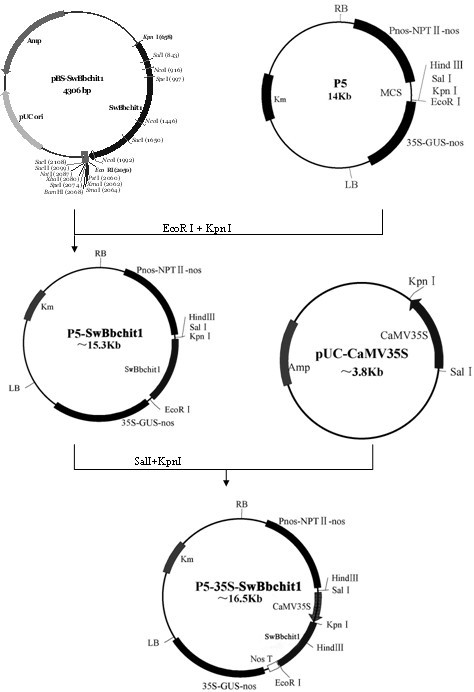
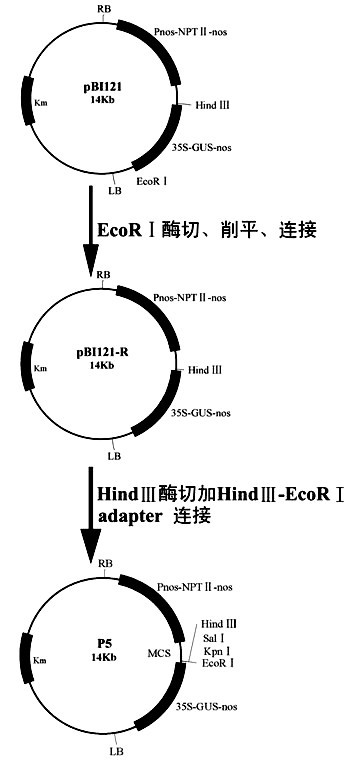
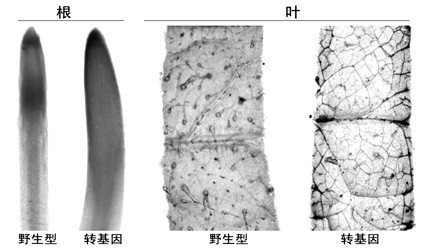
Method for improving beauveria bassiana chitinase gene disease resistance and culturing disease resistance plants adopting method
A technology of Beauveria bassiana and chitinase, which is applied in the field of molecular biology and plant genetic engineering, achieves huge economic and social benefits, remarkable effects, and the effect of improving resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Extraction of DNA
[0056] 1.1 DNA extraction buffer
[0057] (1) Fungal DNA extraction buffer
[0058] Tris-HCl (pH 7.5) 0.2 mol / L, NaCl 0.5 mol / L, EDTA 0.01 mol / L, SDS 1% (w / v).
[0059] (2) Plant DNA extraction buffer
[0060] CTAB extract: 100 mmol / L Tris-HCl (pH8.0), 20 mmol / L EDTA (pH8.0), 1.5 mol / L NaCl, 2% CTAB (w / v), 4% PVP40 (w / v ) and 2% mercaptoethanol (v / v), PVP and mercaptoethanol were added just before use.
[0061] (3) Alkaline lysis plasmid extraction buffer
[0062] STE: 0.1 mol / L NaCl, 10 mmol / L Tris-HCl (pH 8.0), 1 mmol / L EDTA (pH 8.0).
[0063] Solution I: 50 mmol / L glucose, 25 mmol / L Tris-HCl (pH 8.0), 10 mmol / L EDTA (pH 8.0).
[0064] Solution II: 0.2 mol / L NaOH, 1% (w / v) SDS.
[0065] Solution III: 50 mL 5 mol / L potassium acetate, 11.5 mL glacial acetic acid, 28.5 mL water.
[0066] 1.2 Extraction method of Beauveria bassiana genome DNA
[0067] Collect the Beauveria bassiana bacteria liquid in a 1.5 ml centrifuge ...
Embodiment 2
[0074] Example 2: Extraction of RNA
[0075] 2.1 RNA extraction buffer
[0076] CTAB extraction buffer: 2% CTAB (w / v), 2% polyvinylpyrrolidone PVP40 (w / v), 100mmol / L Tris-HCl (pH8.0, prepared in DEPC-treated water), 25mmol / L EDTA, 0.5 g / L Spermidine, 2.0mol / L NaCl, 2% mercaptoethanol (v / v, add before use).
[0077] SSTE solution: 1mol / L NaCl, 0.5% SDS (w / v), 10mmol / L Tris-HCl (pH8.0), 1.0mmol / L EDTA.
[0078] 2.2 Plant RNA extraction method
[0079] Total RNA was extracted from cotton or tomato tissues by CTAB method. Take about 3g of fresh cotton or tomato tissue, quickly grind it into powder in liquid nitrogen, put it into a 50ml centrifuge tube treated with DEPC water, then add 15ml of RNA extraction solution preheated at 65°C, mix it upside down, and then bathe in water at 65°C for 3 minutes. Centrifuge at 8,000 rpm and 4°C for 10 min, transfer the supernatant to a new 50ml centrifuge tube treated with DEPC water, and extract twice with an equal volume of chlorof...
Embodiment 3
[0082] Example 3 Obtaining the chitin-binding domain (SwChBD) of the silkworm chitinase gene
[0083] RNA was extracted from the molting stage of the 4th to 5th instar silkworm (Bombyx mori), and cDNA was synthesized by reverse transcription (product of TaKaRa Company) according to the instructions of the reverse transcription kit. According to the cDNA sequence of the silkworm chitinase gene (Genebank accession number: AF273695), primer sequence 3 and sequence 4 were designed to amplify the chitin-binding domain of the silkworm chitinase gene and introduce Speech I restriction site, C-terminal introduction EcoR I restriction site.
[0084] Components of the amplification reaction: 10×DNA polymerase Buffer (product of Dingguo Company) 2.5 μL, 10 mmol / L dNTP 0.5 μL, 5 μmol / L upstream and downstream primers 1 μL each, DNA polymerase 0.7U, cDNA 1 μL, add water to 25 μL . Cycle conditions: 95°C, 5min; 94°C, 1min; 55°C, 1min; 72°C, 1min, 32 cycles; 72°C, 20min, agarose gel e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com