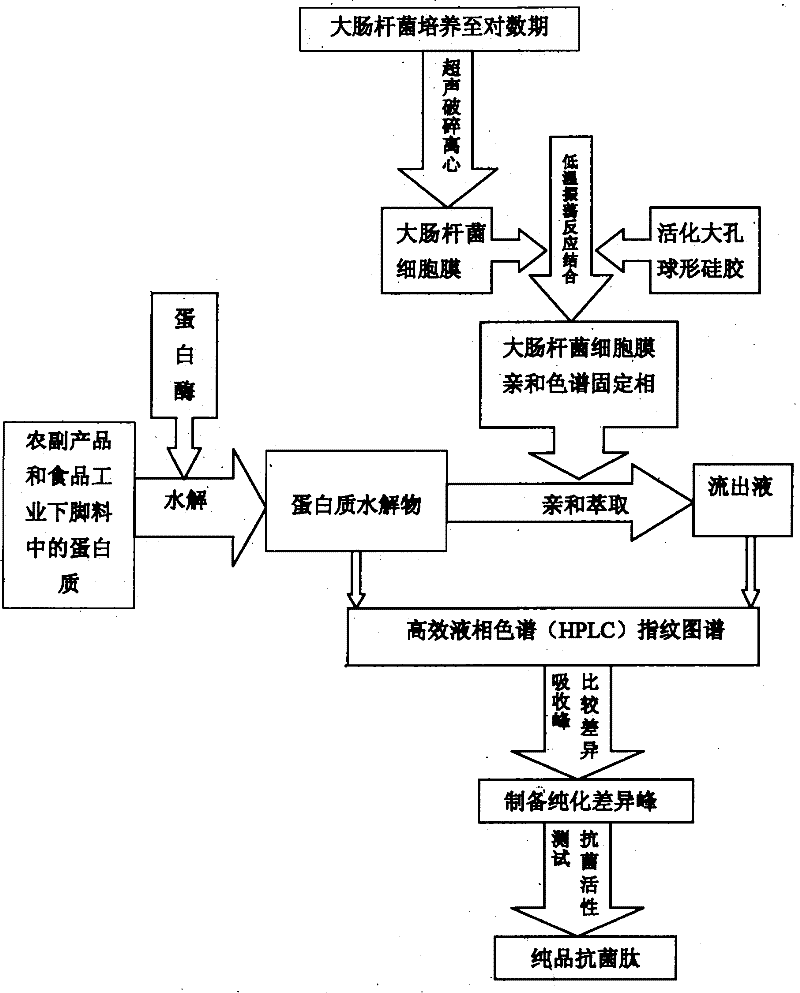
Method for quickly and accurately discovering, identifying and preparing proteolysis provenance antibacterial peptide
A protein hydrolyzate and antimicrobial peptide technology, which is applied in the field of preparation and purification of bioactive peptides, can solve the problems of industrialization difficulties, high cost of chemical synthesis, and high cost, and achieve the effect of reducing production costs and reducing environmental pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Preparation of Affinity Cell Membrane Stationary Phase
[0024] 500 ml of the E. coli culture cultured to the logarithmic phase was centrifuged at 4000 r / min for 15 min to obtain the bacteria, which were washed 10 times with 25 ml of Tris-EDTA buffer (pH 7.4) to remove medium residues. The cleaned cells were reconstituted with Tris-EDTA buffer, and frozen at -30°C for 4h, then removed and thawed in a 37°C water bath. After repeated freezing and thawing 5 times, centrifuged at 3000r / min for 15min. The frozen and thawed bacteria were subjected to cell lysis treatment at 600W ultrasonic power, and the ultrasonic wave was irradiated for 4 seconds each time, with an interval of 4 seconds, and a total time of 60 minutes. The precipitate was obtained by low-speed centrifugation. The precipitate was the cell membrane of E. coli. 10ml of suspension prepared by reconstituted cell membrane was placed in a centrifuge tube, then 0.5g (3-7μm) of activated macroporous spherical...
Embodiment 2
[0025] Example 2 Preparation of protein antibacterial hydrolysate
[0026] Taking Jatropha curcas seed meal protein as an example, take a protein sample of 10 g, with a purity of 92.26%, add 100 mL of deionized water, and use different hydrolytic proteases for hydrolysis. The optimal conditions for protease hydrolysis are shown in Table 1. After the protein solution is hydrolyzed by different proteases to different degrees of hydrolysis (7%, 9%, 11%, 13%, 15% and 17%), the turbid solution is adjusted to pH 7.5 at 45°C, and water bathed at 100°C. Enzyme for 10 minutes, then centrifuged at 4000r / min for 15 minutes, the supernatant was obtained, lyophilized, and reconstituted into a 1mg / ml sample solution, and the antibacterial ability test was performed respectively to find the component with the strongest antibacterial ability and cool to dry. Name this sample Fh13.
[0027] Table 1 Optimal working conditions of protease
[0028]
Embodiment 3
[0029] Example 3 Affinity extraction combined with high performance liquid phase and mass spectrometry technology to accurately and quickly discover and identify antimicrobial peptides
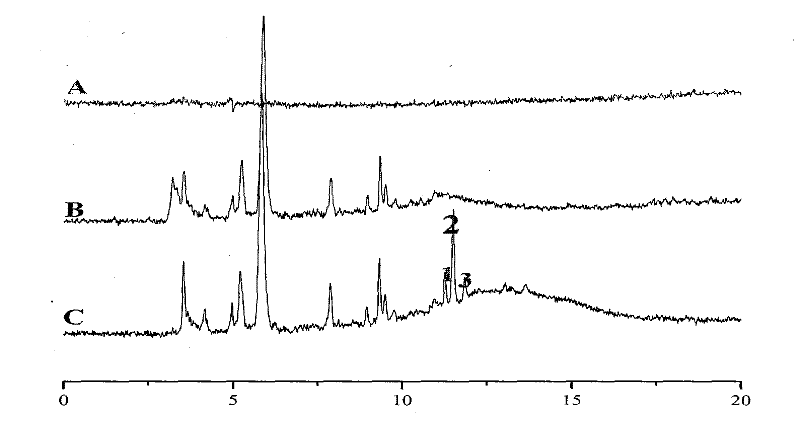
[0030] Place the affinity stationary phase of Example 1 and the medium Fh13 of Example 2 in a 10 ml centrifuge tube, shake and combine at 37° C. for 1 hour, and centrifuge the supernatant. The precipitation was washed 7 times with buffer solution, the washing solution and supernatant were combined, cooled to dryness and named Fh13-1. Reconstitute Fh13 and Fh13-1, and use high performance liquid-mass spectrometry to detect their fingerprints and analyze the difference peaks (as shown in the accompanying drawings image 3 Peaks 1, 2 and 3), prepare the difference peaks and test the antibacterial ability. Under these conditions, it was quickly discovered that a protein hydrolysate-derived antibacterial peptide was prepared. The series of this peptide is CAILTHKR, and the minimum inhibitory solubilit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com