Pyrene-marked single-chain DNA (deoxyribonucleic acid) fluorescent probe and preparation method thereof
A technology of labeling and pyrene, which is applied in DNA preparation, recombinant DNA technology, DNA/RNA fragments, etc. It can solve the problems of complex synthesis operation, and achieve the effect of not losing sensitivity, good reference value and high accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
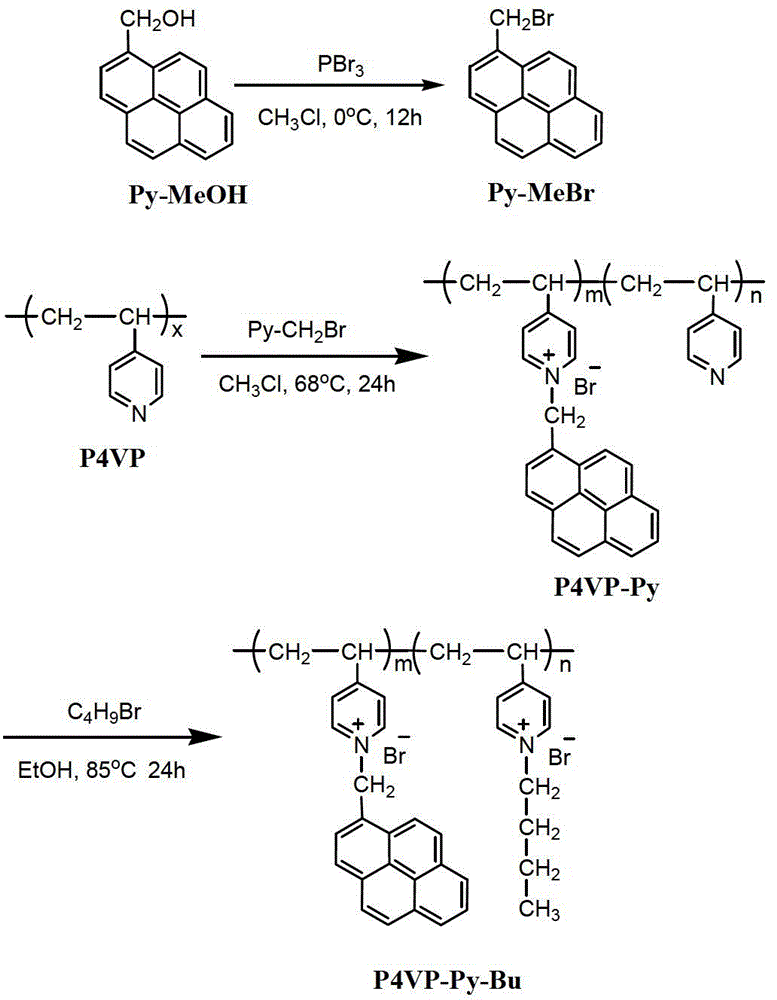
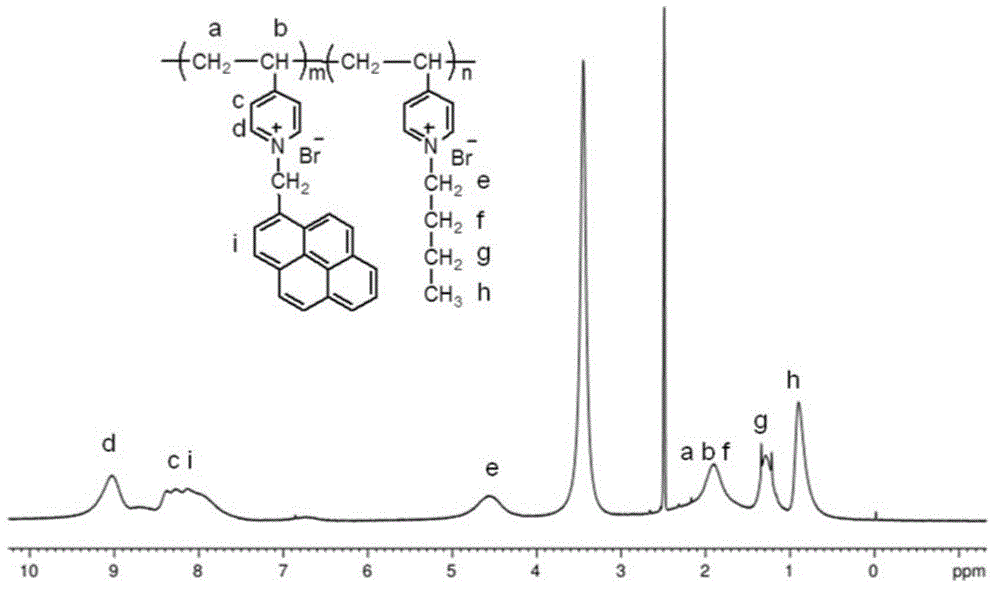
[0044]Dissolve 6 mmol of pyrenemethanol in 150 mL of chloroform at freezing point temperature, add 3 mmol of phosphorus tribromide to the solution, stir in an ice bath for 12 hours, then add saturated aqueous sodium bicarbonate to neutrality, separate layers, and take the organic layer for Distillation, recrystallization, and drying give pyrene bromide. Dissolve 12mmol of poly-4-vinylpyridine in 10mL of chloroform, then add 1.2mmol of pyrenemethyl bromide, stir and reflux at 68°C for 24 hours, then slowly add the reaction solution dropwise into tetrahydrofuran to obtain a precipitate, filter, wash, and dry Then intermediate products are obtained. Dissolve 5 mmol of the intermediate product in 8 mL of ethanol, add 14 mol of n-bromobutane, stir and reflux at 85°C for 24 hours, then slowly add the reaction solution dropwise into tetrahydrofuran to obtain a precipitate, filter, wash, and dry to obtain the product cationic poly Electrolyte (P4VP-Py-Bu).
Embodiment 2
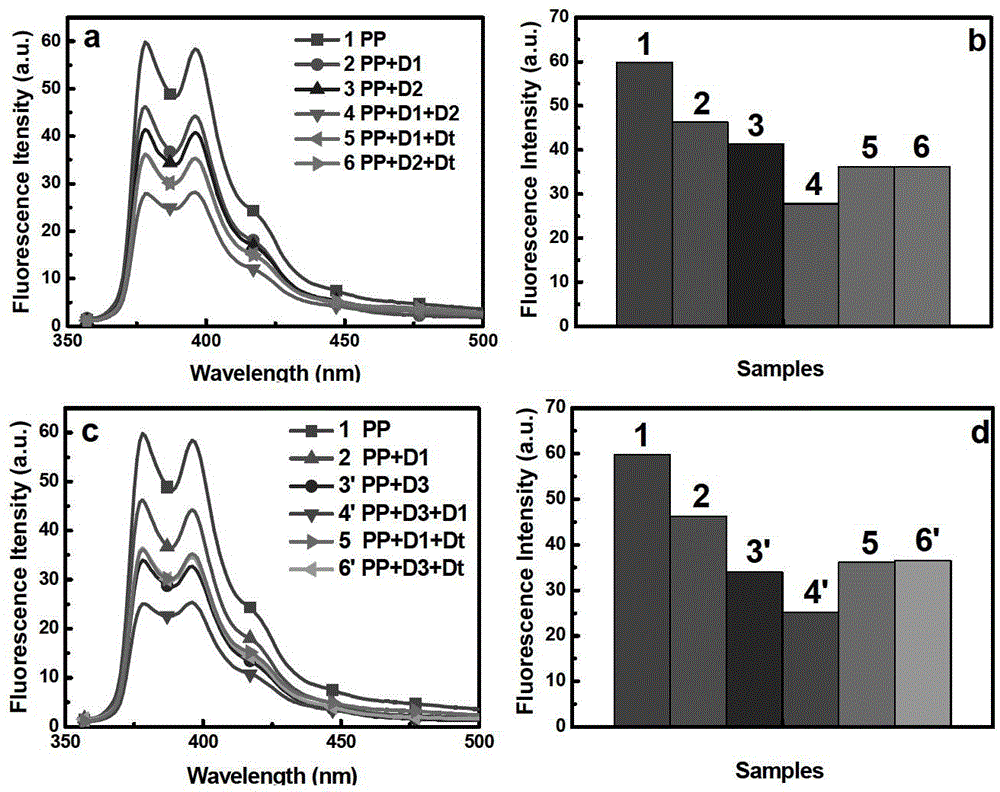
[0046] (1) Preparation of samples: Use PBS (10 mM, pH = 7.4) buffer to prepare each sample solution, as follows: image 3 1) P4VP-Py-Bu, 2) P4VP-Py-Bu / ssDNA1, 3) P4VP-Py-Bu / ssDNA2, 4) P4VP-Py-Bu / ssDNA1+ssDNA2, 5) P4VP-Py- Bu / ssDNA1+ssDNAt, 6) P4VP-Py-Bu / ssDNA2+ssDNAt, 3') P4VP-Py-Bu / ssDNA3, 4') P4VP-Py-Bu / ssDNA2+ssDNA3, 6') P4VP-Py-Bu / ssDNA3+ssDNAt total 9 samples, the final concentration of polyelectrolyte in each sample solution was 2.0×10 -7 M, each DNA concentration is 1.0×10 -6 M.
[0047] (2) Heat treatment sample: Put the sample prepared in (1) into a 90°C oven for heat treatment for 15 minutes, and then quickly move to a 40°C oven for heat treatment for 30 minutes.
[0048] (3) Fluorescence detection of the sample: under the condition of excitation wavelength of 344nm, pass image 3 It can be clearly seen from the graph and histogram of fluorescence intensity at 377nm that when DNA1 or DNA3 interacts with polyelectrolyte to form a fluorescent probe, the fluore...
Embodiment 3
[0050] (1) Preparation of samples: Use PBS (10 mM, pH = 7.4) buffer to prepare each sample solution, as follows: Figure 5 1) DNA1 / DNA2, 2) P4VP-Py-Bu + DNA1 / DNA2, 1’) DNA1 / DNA3, 2) P4VP-Py-Bu + DNA1 / DNA3 marked in a total of 4 samples. The final concentration of polyelectrolyte in each sample solution was 2.0 × 10 -5 M, each DNA concentration is 1.0×10 -4 M.
[0051] (2) Heat treatment sample: Put the sample prepared in (1) into a 90°C oven for heat treatment for 15 minutes, and then quickly move to a 40°C oven for heat treatment for 30 minutes.
[0052] (3) Circular dichroism test on the sample: The circular dichroism test is done to confirm that the pyrene molecule is indeed intercalated into the DNA double helix structure. Such as Figure 5 In the spectrum shown, the circular dichroism intensity after adding polyelectrolyte P4VP-Py-Bu all decreased significantly, and the positive band appeared blue shifted, and the negative band appeared red shifted. The only possibili...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com