Amphioxus agglutinoid and expressed genes and application thereof
A lectin-like, gene expression technology, applied in the field of genetic engineering, can solve problems such as hindering the rapid development of marine aquaculture
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Cloning of the gene encoding the lectin AmphiITLN239631 of Wenchang fish in Qingdao:
[0032] 1.1 For the extraction of Qingdao amphioxus total RNA and the synthesis of first-strand cDNA, the extraction and synthesis steps refer to the relevant kit instructions of Takara and TIANGEN companies.
[0033] 1) Cut 100mg of fresh tissue (adult amphioxus) as much as possible into a pre-1.5mlep tube.
[0034] 2) Add 1ml Trizol (Invitrogen), quickly grind the tissue with a grinding pestle until there are no large tissue pieces, and let stand at room temperature for 5 minutes to fully lyse the tissue. At this time, the samples can be stored at -80°C for long-term storage.
[0035] 3) Add 0.2ml of chloroform (chloroform) to every 1ml of Trizol, cover and seal tightly, shake vigorously for 15s, and let stand at room temperature for 15 minutes.
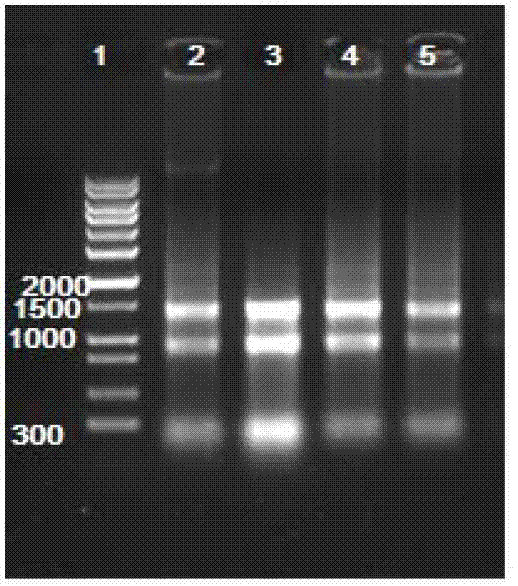
[0036] 4) Centrifuge at 1000g for 15min at 4°C. At this time, the sample is divided into three layers: the red organic phase of the low...
Embodiment 2
[0056] Gene sequence determination
[0057] 2.1. Ligation of DNA fragments to cloning vectors
[0058] The recovered DNA fragments were connected to the pEASY-T3 vector of Transgene Company. Ligation reaction system: 1 μl of ligation carrier enzyme mixture, 4 μl of foreign DNA fragments, flick the centrifuge tube with your finger, mix the sample evenly, turn it on the centrifuge for 2 seconds, concentrate the sample at the bottom of the tube, and ligate at 37°C for 5 minutes.
[0059] 2.2. Prepare Escherichia coli DH5α competent according to the method for preparing Escherichia coli competent in "Molecular Cloning Experiment Guide".
[0060] 2.3. Transfer the ligated recombinant vector pEASY-T3 to Escherichia coli DH5α competent according to the heat shock transformation method in the "Molecular Cloning Experiment Guide".
[0061] 2.4. The transformed Escherichia coli DH5α was spread on MacConkey medium containing 100 μg / L ampicillin, cultured overnight at 37°C, and the ...
Embodiment 3
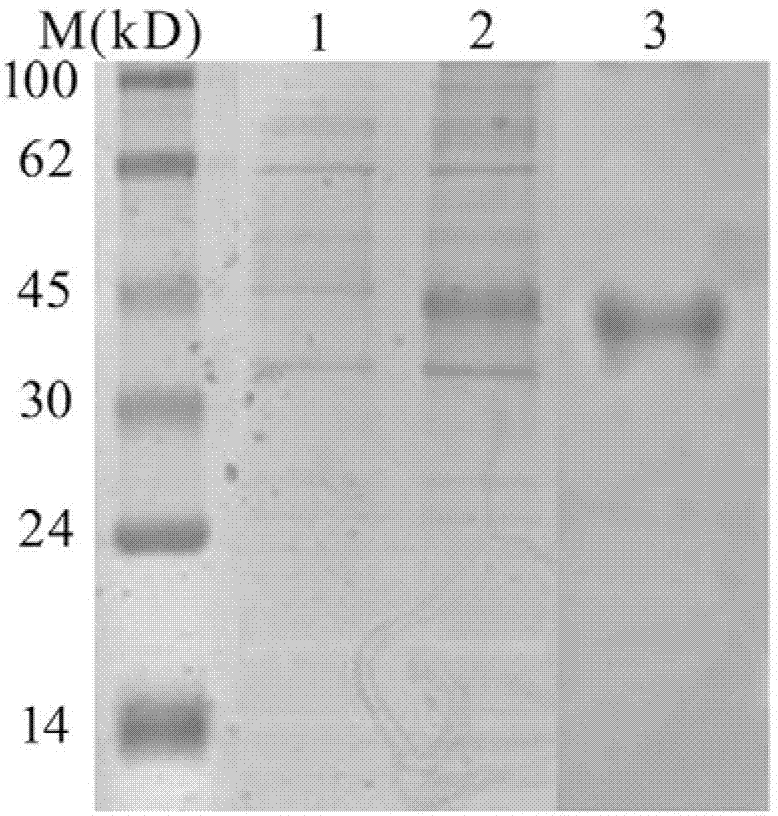
[0066] Heterologous Expression and Purification of Lectin-like AmphiITLN239631
[0067] 3.1 Construction of expression vector pET29b-AmphiITLN239631
[0068] 1) Using the small-dose plasmid extraction kit, follow the steps in the instructions to extract the recombinant plasmid pEASY-T3-AmphiITLN239631 from the transformant containing the recombinant plasmid pEASY-T3.
[0069] 2) Design two specific primers according to the sequencing results:
[0070] pET29b-Amphi239631-F: 5'GAATTCCATATGACGCCCGAGG 3' and
[0071] pET29b-Amphi239631-R: 5' CCGCTCGAGACGGTAAAAGAAG,
[0072] The above primers were synthesized by Shanghai Boshang Biotechnology Company.
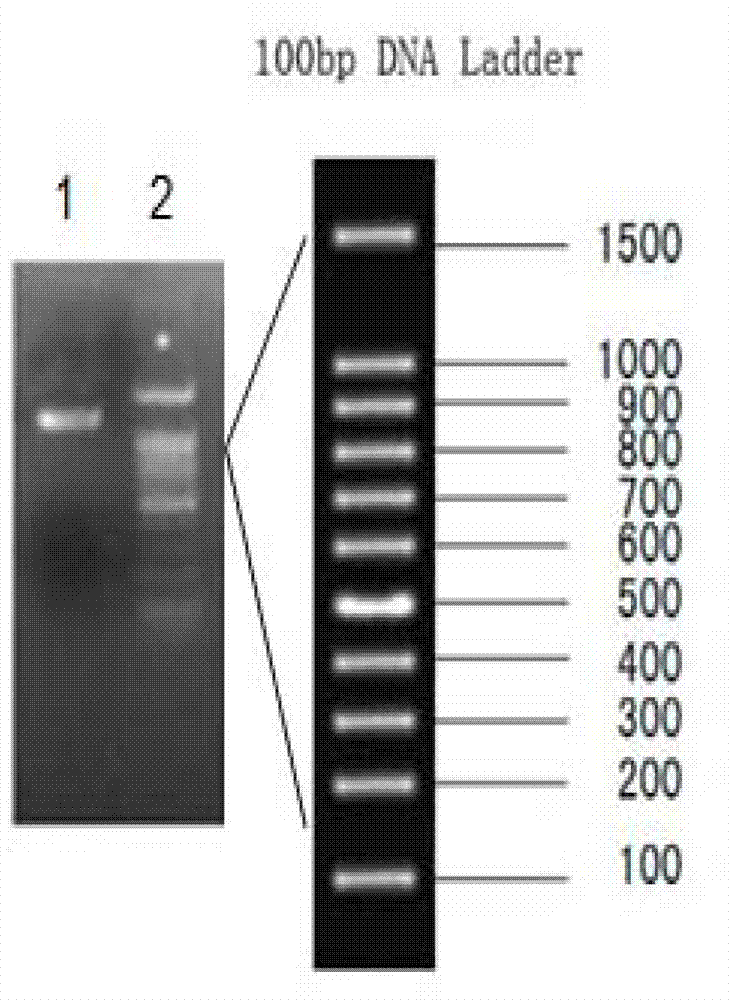
[0073] 3) The gene sequence was amplified by PCR, using pET29bAmphiITLN239631-F and pET29b-AmphiITLN239631-R as primers, and the recombinant plasmid pEASY-T3-AmphiITLN239631 as a template for PCR amplification. The PCR reaction conditions were: 95°C, 5 minutes; 95°C, 1 minute; 55°C, 1 minute; 72°C, 2 minutes, after 35 cycles, 72...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com