Proteusbacillus vulgaris multi-serotype strain specific primer and multiple polymerase chain reaction (PCR) detection method
A proteus, specific technology, applied in the field of multiplex PCR rapid detection, can solve the problems of time-consuming, missed detection, antiserum limitation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0126] Example 1 : Genome Extraction
[0127] Proteus was cultured in a small shaking tube equipped with 2YT, and the cells were collected after overnight culture at 37°C and 180rpm in a shaker, and the genome was extracted by traditional phenol and chloroform extraction method. Specific steps are as follows:
[0128] (1) Collect 1ml of the bacterial liquid into a 1.5ml centrifuge tube, centrifuge at 12000rpm for 10min, discard the supernatant to collect the bacterial cells.
[0129] (2) Add 500 μl tris-Hcl to the centrifuge tube to resuspend the cells, and centrifuge at 12,000 rpm for 5 minutes to remove the supernatant.
[0130] (3) Repeat step (2)
[0131] (4) Add 250ul 50mM Tris-HCl (pH8.0) and 10ul 0.4M EDTA to the cells to resuspend the cells, add 10ul 10mg / ml lysozyme and incubate at 37°C for 20 minutes. For Gram-positive bacteria overnight.
[0132] (5) Add 3ul 20mg / ml proteinase K and 15ul 10% SDS to the mixture, incubate at 50°C for 2 hours. If the liquid does...
Embodiment 2
[0138] Embodiment 2: the design of primer
[0139] Using the Proteus sequence tested by the laboratory, for serotypes 1, 2, 3, 4, 7, 8, 9, 10, 11, 12, 13, 14, 17 Type, Type 18, Type 19, Type 21, Type 23, Type 24, Type 25, Type 26, Type 27, Type 28, Type 29, Type 30, Type 31, Type 32, Type 33, Type 34, Type 36, Type 37, Type 38, Type 39, Type 40, Type 41, Type 44, Type 45, Type 47, Type 48, Type 50, Type 52, Type 53, Type 54, Type 55, Type 56, Type 57, Type 58 , Type 59, and Type 60's wzy Primers were designed for the specific regions of the genes, and the primer sequences are shown in Table 1 below:
[0140] Table 1 Sequences of specific primers for 48 serotypes of Proteus
[0141]
[0142]
[0143]
[0144]
[0145]
Embodiment 3
[0146] Example 3 : Screening of specific primers
[0147] Primers were designed for specific regions of the wzt gene of 48 Proteus serotypes, and the primer sequences are shown in Table 1 (SEQ ID NO:1-SEQ ID NO:96). One standard strain of each of the 48 Proteus serotypes, one strain of each of the 10 standard strains of other Proteus serotypes, and six closely related bacteria were collected to verify the specificity of the primers. The strain numbers and sources are shown in Table 2 below.
[0148] Table 2 Standard strains for testing
[0149]
[0150]
[0151]
[0152]
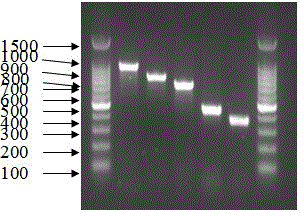
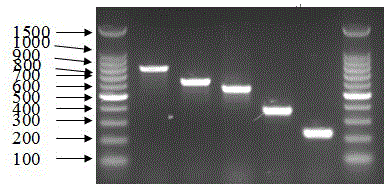
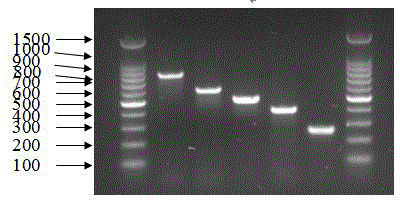
[0153] The PCR system is 10uM primers are added according to the instructions in Table 3, MgCl 2 3μl, 10×buffer 3μl, 10mM dNTP 2.4μl, 5 U / μl Taq polymerase 0.3μl and 2μl sample template to be tested into a 0.2ml thin-walled PCR tube, and finally use ddH 2 O to make up to 30 μl. All primers obtained positive results in the 48 Proteus templates within the detection range, and did not get an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com