Mannase and mutants thereof
A mannanase, mutant technology, applied in the field of microbial engineering, can solve problems such as difference in activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Cloning of Bacillus licheniformis mannanase gene
[0045] 1.1 Extraction of total genomic DNA from different strains of Bacillus licheniformis
[0046] Cultivate the 6 strains of Bacillus licheniformis preserved in our laboratory overnight, take 1.5ml each, centrifuge at 12000rpm for 1 minute, remove the supernatant; add 200μl lysis buffer (60mM Tris-HCl, pH7.8, 20mM Na-Ac, 1mM EDTA , 1.5% SDS), blow vigorously with a pipette; add 66μl 5M sodium perchlorate solution to mix, centrifuge at 12000rpm for 10 minutes, and take the supernatant; add an equal volume of phenol to extract once, centrifuge at 12000rpm for 2 minutes, and take the supernatant; Add an equal volume of isopropanol to precipitate for 5 minutes, centrifuge at 12000rpm for 5 minutes; wash twice with 70% ethanol; finally dissolve the dried DNA in ddH 2 O.
[0048] Using the total genomic DNA extracted in 1.1 as a template, PCR amplification was performed using primer...
Embodiment 2
[0051] Example 2 Sequence Analysis of Mannanase
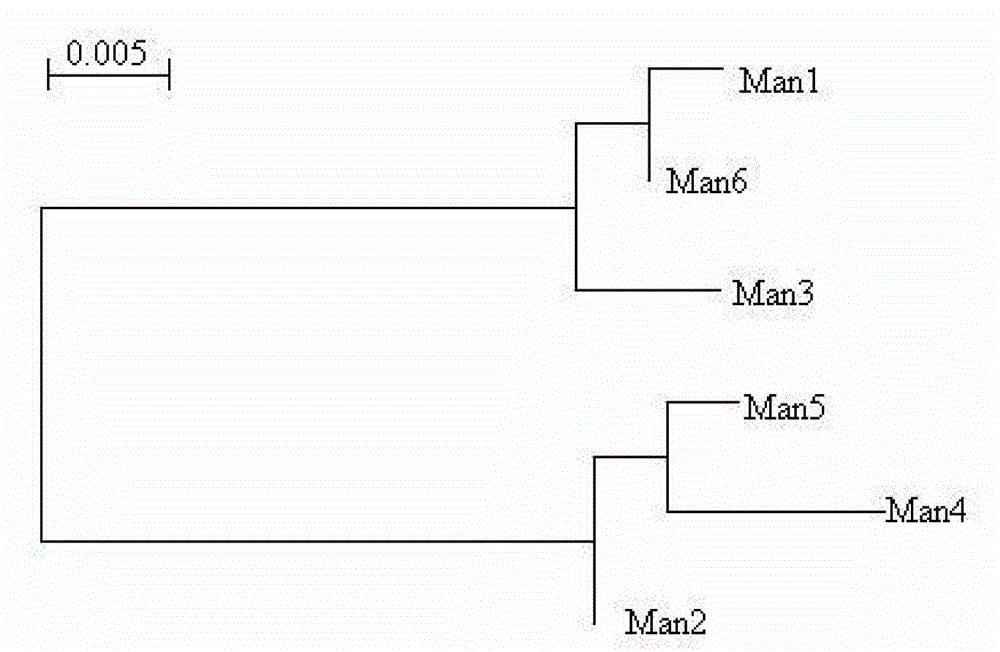
[0052] BLAST analysis of the sequencing results on NCBI showed that these sequences were highly homologous to the mannanase gene. Using the biological software DNAMAN6.0 to analyze the nucleic acid sequences, it was found that the six sequences were not completely homologous. The corresponding nucleic acid sequences were translated into amino acid sequences and named as Man1, Man2, Man3, Man4, Man5 and Man6, respectively. The amino acid sequence homologous comparison analysis shows that there are mutations at one or more of the following positions, namely: 20, 27, 31, 48, 64, 78, 81, 89, 102, 105, 122, 123, 179 , 187, 195, 234, 248, 254, 255, 292, 302, 312, 313 and 325 positions. Possible substitutions for amino acid residues at these positions are Y20N, D27N, N31 S, L48 T, V64I, E78 K, V81D, S89R, R102Q, L105M, S122P, S123N, S179T, A187E, R195Q, H234Y, H248Y, D254E, Q255E, N292K, G302E, G312D, A313T, D325E, the results of...
Embodiment 3
[0053] Example 3 Recombinant expression and purification of mannanase in Escherichia coli
[0054] Using the plasmids pMDT-Man1, pMDT-Man2, pMDT-Man3, pMDT-Man4, pMDT-Man5 and pMDT-Man6 as templates, primers (AGA GCTAGC GCACACACCGTTTCTCCGGTG --- Nhe I and ACA CTCGAG CACGACAGGCGTCAAAAGAATCG--- xho I) Carry out PCR amplification. The PCR amplification conditions are 95°C for 4min; 94°C for 30S; 55°C for 40S; 72°C for 1min for 30 cycles; 72°C for 7min. After the gel recovery of the amplification products, the first Nhe I digest, and then recover the digested product to carry out xho I digestion. Similarly, the expression plasmid pET28a was also carried out separately Nhe I digestion and xho I digestion. The cloned gene and the expression vector were ligated overnight at 4°C with T4 ligase. Finally, the ligated product was introduced into E. coli BL21. The expression plasmids of the corresponding positive clones were named pET-Man1, pET-Man2, pET-Man3, pET-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com