Establishment method of streptomyces diastatochromogenes expression system
A Streptomyces chromogenes and expression system technology, applied in the field of genetic engineering, can solve the problems of low transcriptional activity, ineffectiveness, and lack of versatility of promoters
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
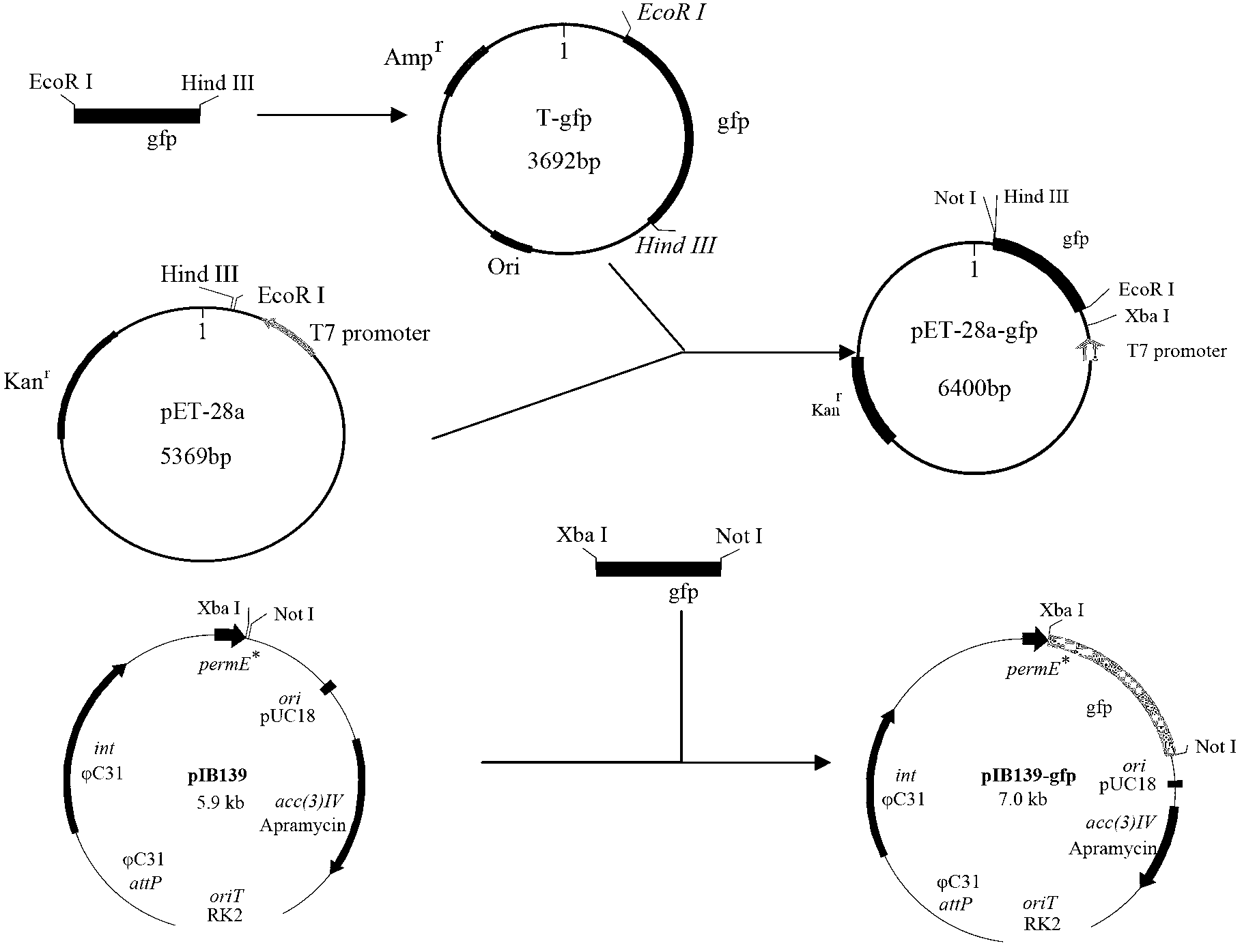
[0035] Example 1: Using the green fluorescent protein gene gfp Construction of the genetic expression system of amylase Streptomyces chromogenes as a reporter gene:
[0036] Using plasmid pCAMBIA1302 as template, P gfp f Eco R I and P gfp R Hin dIII is the primer PCR obtained containing Eco R I and Hin dIII two enzyme cutting sites gfp Fragment, connected with pMD18-T Vector to construct cloning vector pMD18-T- gfp ( Eco R I+ Hin dIII). The cloning vector pMD18-T- gfp ( Eco R I+ Hin dIII) converted to E. coli JM109 recipient bacteria were spread on the LB agar plate containing Amp, IPTG, and X-gal. After culturing overnight at 37 °C, many white colonies grew on the plate. Randomly pick white clones for enzyme digestion identification and send them to Shanghai Sangon Bioengineering Co., Ltd. for sequencing Eco R I and HindIII double digestion to obtain Eco R I and Hind III of the two enzyme cleavage sites gfp The fragment was ligated wit...
Embodiment 2
[0042] Example 2: Using the green fluorescent protein gene gfp Preliminary evaluation of the genetic expression system of amylase Streptomyces chromogenes as a reporter gene:
[0043] Pick the colonies of Streptomyces amylase chromogenes grown on the plate containing 50 μg / mL apramycin to inoculate the CP medium, shake overnight at 28 °C and 180 r / min, and then inoculate them in the Fill with 50 mL of fresh CP medium, culture to mid-logarithmic growth, dry cell weight (DCW) is about 0.3 g / L, collect mycelium by centrifugation at 6000 r / min, wash mycelium twice with PBS buffer Afterwards, resuspend in 30 mL PBS. Take 5-10 μL bacterial liquid smear, observe under Olympus BX50 fluorescence microscope, and take pictures with Color Video Camera of SONY 3CCD at the same time. Using wild bacteria as a control, select the excitation filter to be 485 nm and the emission filter to be 520 nm to determine the transcriptional activity of the promoter.
[0044] The results showed that t...
Embodiment 3
[0045] Example 3: Vitella hyaline hemoglobin gene vgb Expression in amylase Streptomyces chromogenes:
[0046] recombinant plasmid pIB139- vgb builds like Figure 6 As shown, the steps are related to the recombinant plasmid pIB139- vgb The method of construction is similar.
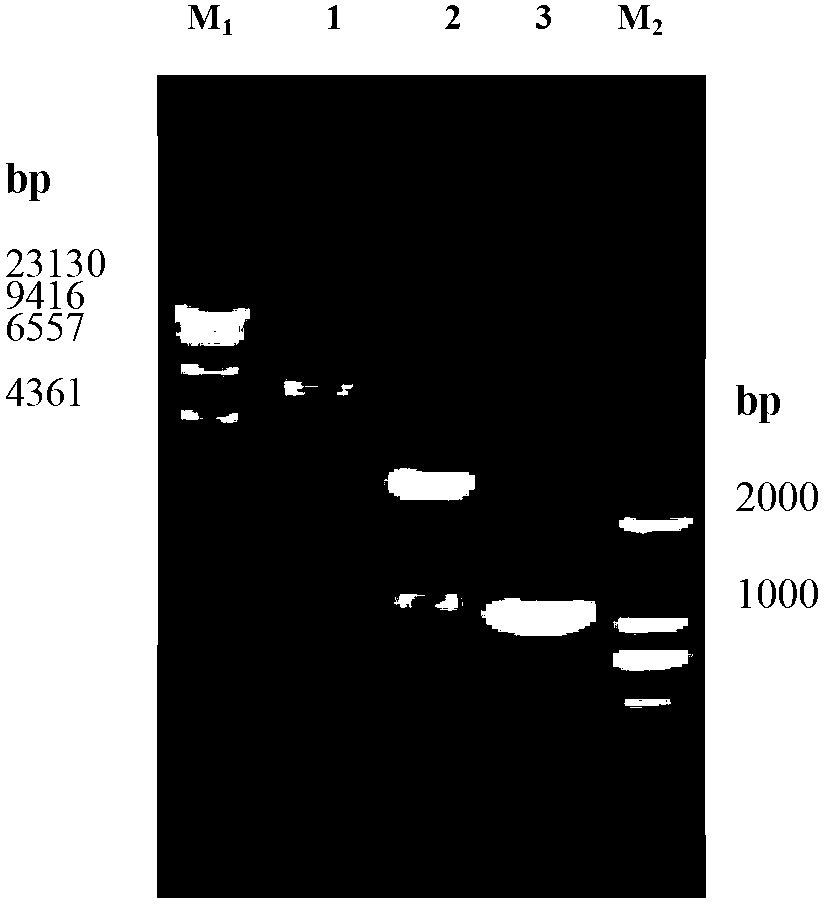
[0047] like Figure 7 and Figure 8 Shown, respectively, recombinant plasmid pET28a- vgb and pIB139- vgb Enzyme digestion verification results. Recombinant plasmid pET28a- vgb through Eco R I and Hin DNA fragments of about 0.5kb and 5.3kb were obtained after dIII double digestion, respectively vgb The size of the gene fragment is the same as that of the plasmid pET28a. recombinant plasmid pIB139- vgb through Nde I and not I obtained a DNA fragment of about 0.6kb after double enzyme digestion, and vgb Gene fragments are of the same size. The above results prove that the recombinant plasmid pIB139- vgb builds correctly.
[0048] Extract plasmid pIB139- vgb , into amylase S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com