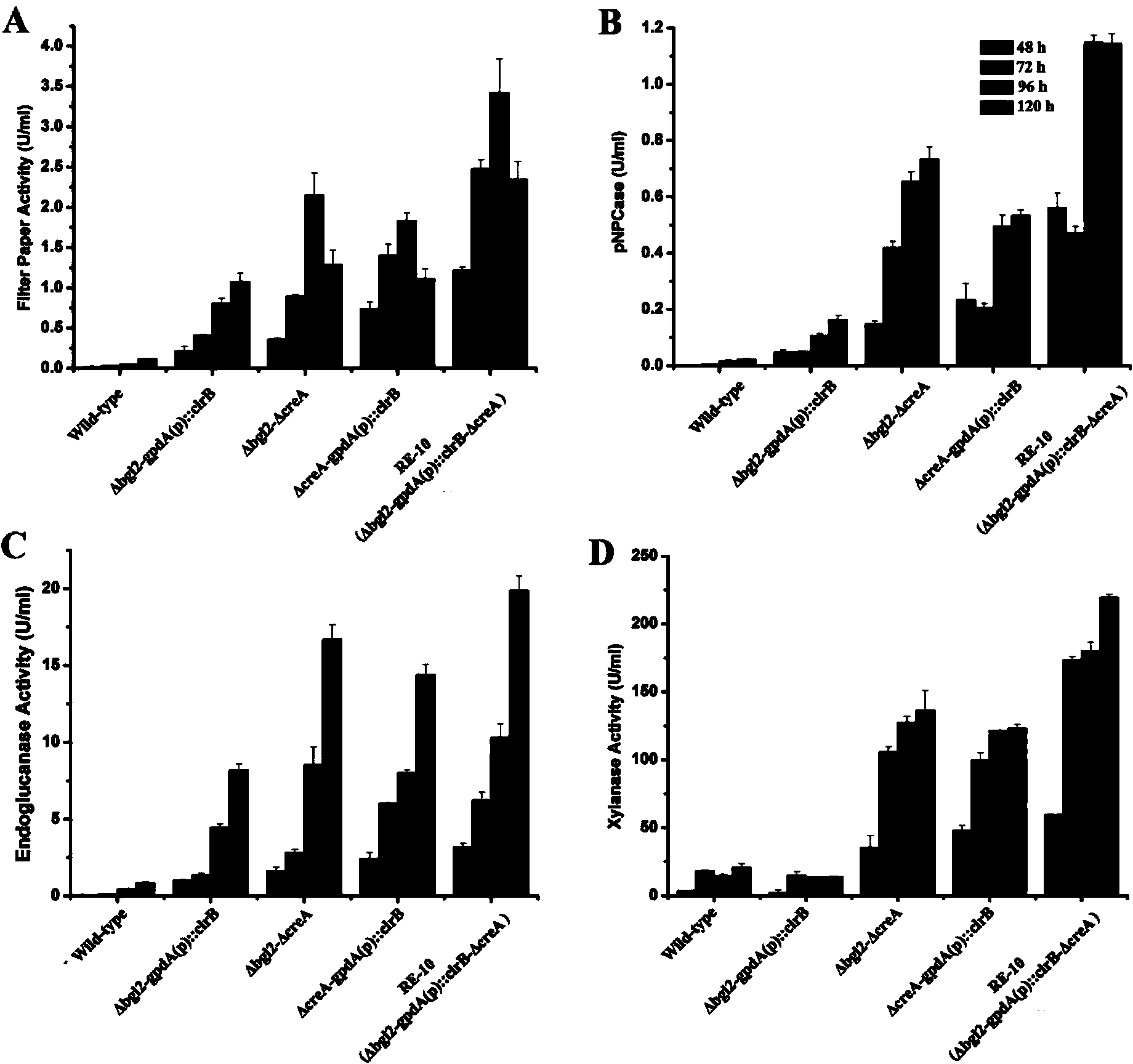
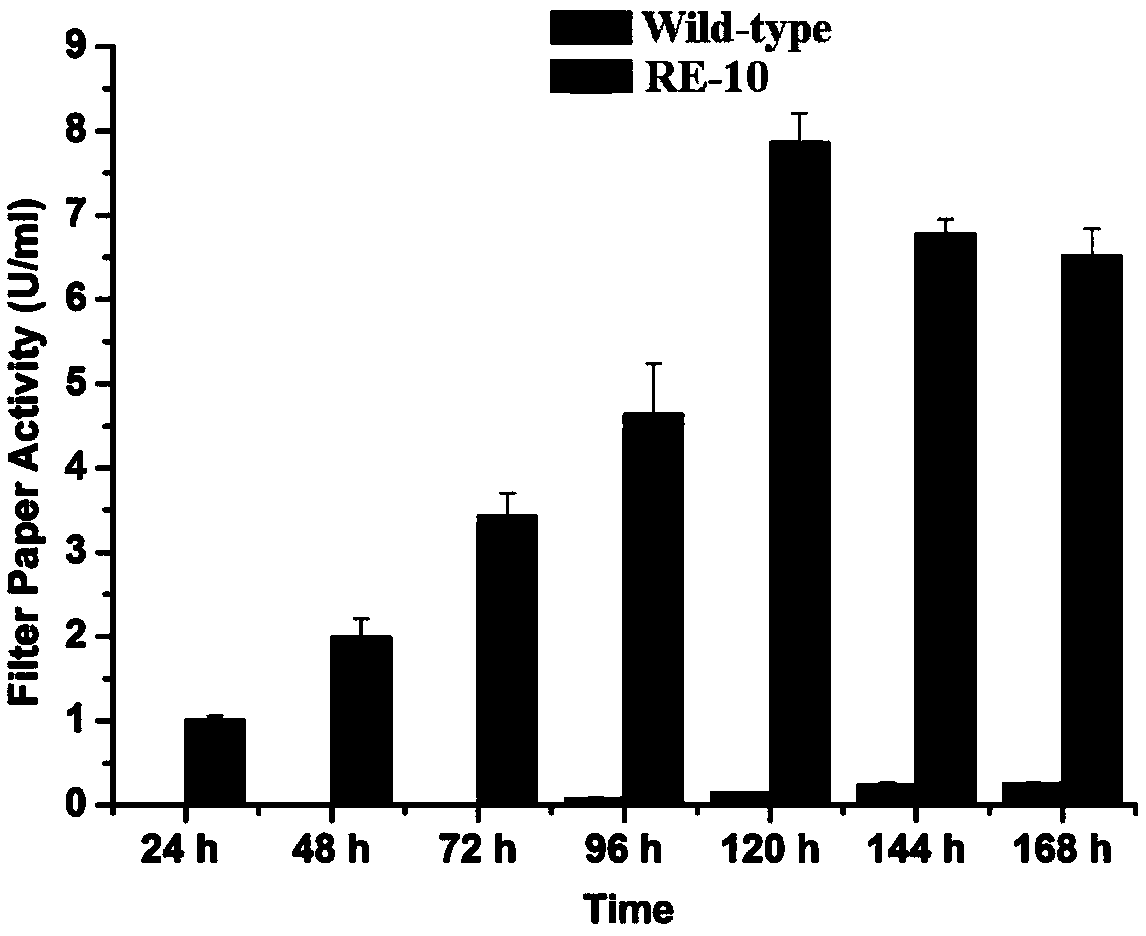
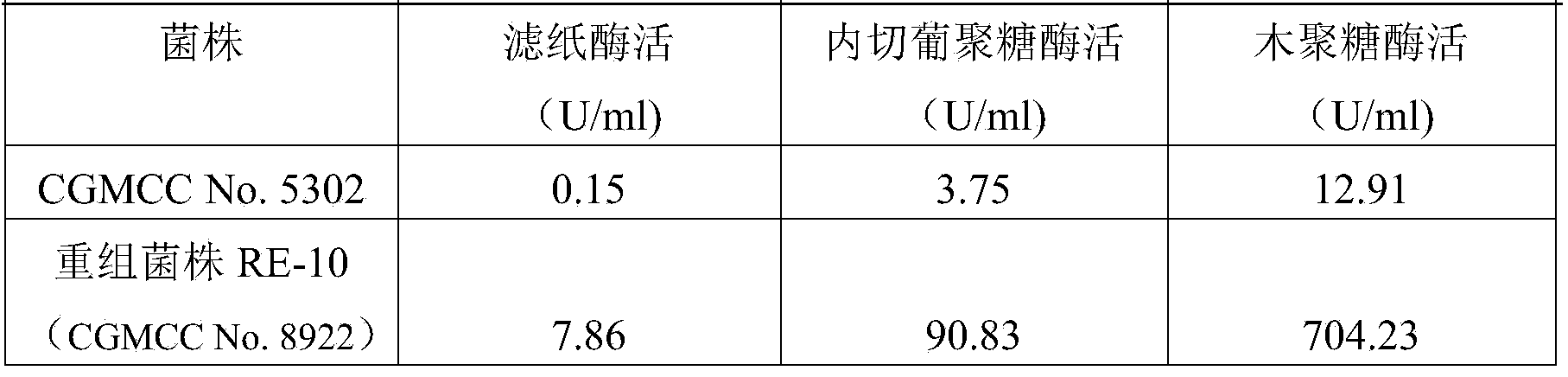
Penicillium oxalicum strain for increasing enzyme activities of cellulase and hemicellulase
A hemicellulase enzyme, Penicillium oxalicum technology, applied in the field of genetic engineering, can solve the problems of genetic modification without high-yielding cellulase strains, cell function differences, delayed expression, etc., and achieve good industrial development and application prospects and production capacity The effect of strengthening and shortening the production cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Embodiment 1, the construction of Penicillium oxalicum ClrB gene overexpression strain
[0069] (1) Cloning of Penicillium oxalicum ClrB coding region and terminal subregion
[0070] Using the extracted Penicillium oxalicum genomic DNA as a template, specific primers clrB-Fa and clrB-Ra were designed in the ClrB coding region and terminal subregion for PCR amplification to obtain Fragment 1. The primer sequences are as follows:
[0071] upstream primer
[0072] clrB-Fa:
[0073] 5'-AACAGCTACCCCGCTTGAGCAGACATCACCATGTTCCACACCTTTGAAGG-3'
[0074] downstream primer
[0075] clrB-Ra:
[0076] 5'-CCAATGGGATCCCGTAATCAATTGCCCGTAGCACCAGCGAAACATAC-3'.
[0077] (2) Penicillium oxalicum ClrB overexpression cassette promoter region gpdA (p) clone
[0078] The ClrB overexpression box promoter adopts the promoter sequence derived from the gene encoding 3'-phosphoglyceraldehyde dehydrogenase (gpdA) of Aspergillus nidulans. Amplification of the gpdA promoter sequence in a plasmi...
Embodiment 2
[0104] Embodiment 2, knockout of Penicillium oxalicum CreA coding gene
[0105] (1) Cloning of the upstream homology arm fragment of Penicillium oxalicum ⊿creA::bar knockout cassette
[0106] Using the extracted Penicillium oxalicum genomic DNA as a template, design specific primers CreA-F2 and CreAbar-R for PCR amplification of fragment 1 of the upstream homology arm of the CreA-encoded gene knockout cassette. The primer sequences are as follows:
[0107] Upstream primer CreA-F1:5'-CATTCCAGAGATGAACGACC-3'
[0108] downstream primer
[0109] CreAbar-R: 5'-GAAGGCCTGTAAGCGAATTAGCAAGCGTCGATGTGGGAACACCGGATA T-3'.
[0110] (2) Cloning of the homology arm fragment downstream of the Penicillium oxalicum ⊿creA::bar knockout cassette
[0111] Using the Genomic DNA of Penicillium oxalicum extracted by the method in the above general description as a template, design specific primers CreAbar-F and CreA-R2 for PCR amplification of the downstream homology arm fragment 2 of the knockout ...
Embodiment 3
[0130] Embodiment 3, knockout of Penicillium oxalicum β-glucosidase Bgl2 coding gene
[0131] (1) Cloning of the homology arm fragment upstream of the Penicillium oxalicum ⊿bgl2::hph knockout cassette
[0132] Using the extracted Penicillium oxalicum genomic DNA as a template, design specific primers bgl2-F2 and bgl2Hph-R for PCR amplification of fragment 1 of the upstream homology arm of the Bgl2 coding gene knockout cassette. The primer sequences are as follows:
[0133] Upstream primer Bgl2-F1:5'-GGCGGATACCCACTATGACC-3'
[0134] Downstream primer Bgl2hph-R:
[0135] 5'-GCTCCTTCAATATCAGTTAACGTCGTGGGTGTTGGTGGCAGAGTG-3'.
[0136] (2) Cloning of the downstream homology arm fragment of Penicillium oxalicum Bgl2 knockout cassette
[0137] Using the extracted Penicillium oxalicum genomic DNA as a template, design specific primers bgl2Hph-F and bgl2-R2, and PCR amplify the downstream homology arm fragment 2 of the Bgl2 coding gene knockout cassette. The primer sequences are as f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com