High-expression water-soluble heparinase I fusion protein and coding gene thereof
A technology of fusion protein and coding gene, applied in the field of genetic engineering, can solve problems such as poor water solubility, low activity, and poor water solubility of recombinant enzymes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1, expression of heparanase I fusion protein pColdTF-HepI
[0027] 1. Cloning of Flavobacterium heparinase I coding sequence with signal peptide removed
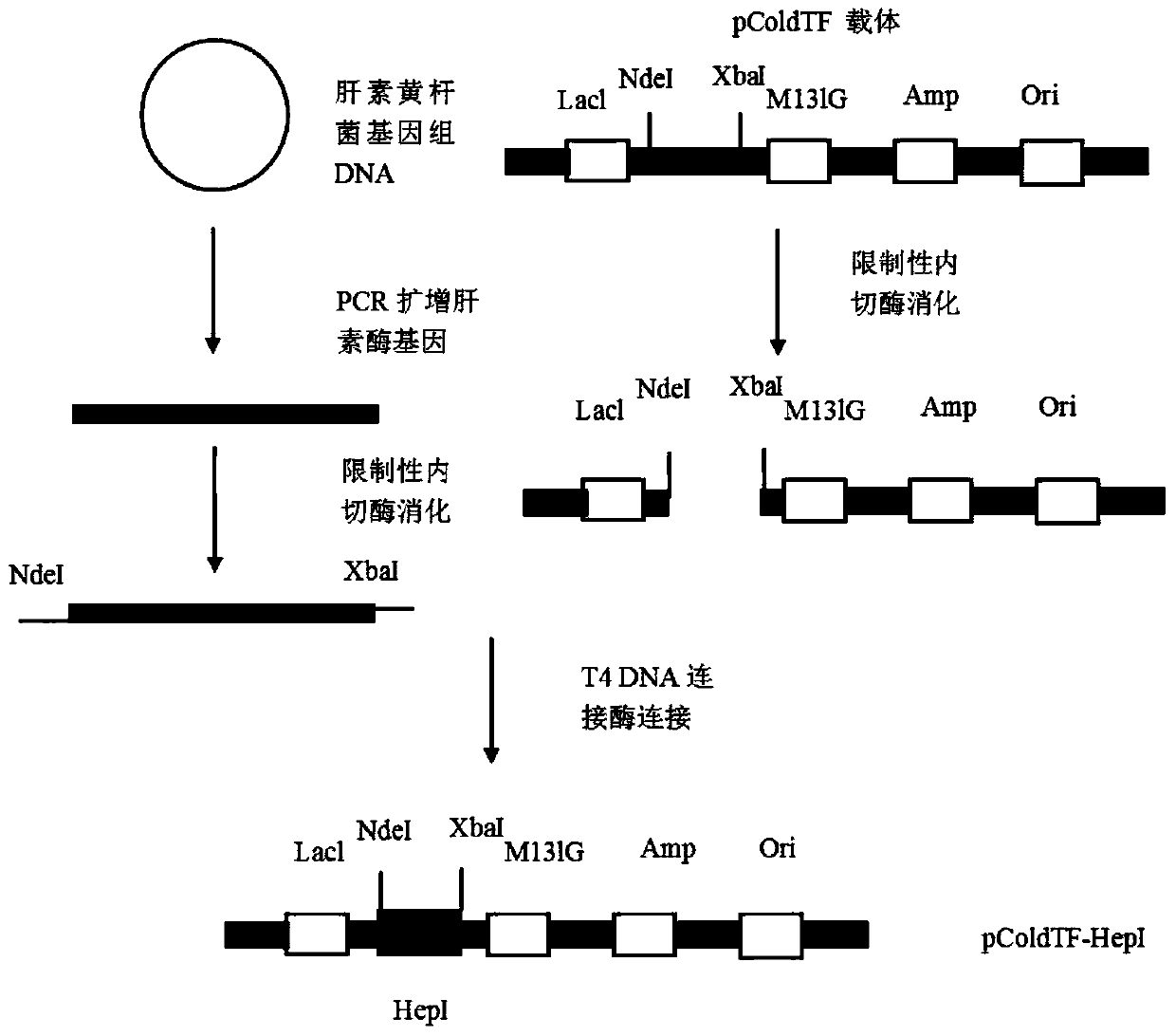
[0028] The construction process of the expression vector pColdTF-HepI is as follows: figure 1 As shown, the specific process is as follows:
[0029] 1. Design and synthesis of primers
[0030] The DNA sequence of heparinase I from Flavobacterium heparinum (Su, H., Blain, F., Musil, R.A., Zimmermann, J.J., Gu, K. and Bennett, D.C. coli of hepB and hepC, genes coding for the glycosaminoglycan-degrading enzymes heparinase II and heparinase III, respectively, from Flavobacterium heparinum.Appl.Environ.Microbiol.1996, 62, 2723-2734), and then according to the removal of heparin encoding signal peptide base The DNA sequence design primer of Flavobacterium heparanase I, and introduce the recognition site of restriction endonuclease xba I and Nde I in primer sequence, used upstream and downstream primers are res...
Embodiment 2
[0056] Example 2, Purification of heparanase I fusion protein pColdTF-HepI by nickel column
[0057] pColdTF-HepI was transformed into Escherichia coli strain BL21(DE3) (purchased from Novagen, USA), and then induced expression with recombinase according to the operation steps provided by the company. And the HepI fusion protein was purified with Ni Sepharose6Fast Flow (GE) gel, and the purification conditions were operated according to the product manual of GE Company. The purification of recombinant pColdTF-HepI was detected by polyacrylamide gel electrophoresis, and the results were as follows: Figure 5 As shown in the No. 5 swimming lane of , the purified recombinant HepI fusion protein presents a single band on the electrophoresis gel, and the position coincides with the predicted molecular weight.
Embodiment 3
[0058] Embodiment 3, the enzyme activity assay of HepI fusion protein
[0059] The mass concentration was 1% heparin, pColdTF-HepI enzyme solution, 5 times buffer solution (250mM Tris-HCl, 500mM NaCl, 10mM CaCl 2, pH7.9) and water were mixed in the ratio of 2:1:2:5 (volume ratio), reacted at the optimum temperature and pH for 2-10min, and measured the enzyme activity by the aforementioned ultraviolet method (Yamagata, Saito et al.1968), while using the protein quantification kit purchased from Kangwei Century Company to measure the protein content of the HepI fusion protease solution, the results showed that the specific activity of the recombinant HepI fusion protein to heparin was 200U / mg.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Extinction coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com