Application of gene NgAUREO1 of nannochloropsis gaditana to adjustment and control of metabolism of fatty acid
A technology of Nannochloropsis and genes, which is applied in the fields of application, genetic engineering, and plant genetic improvement, and can solve problems such as genes that have not been proved by research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Cloning of Example 1 NgAUREO1 Gene
[0026] The aureochromel gene fragment was found from the Nannochloropsis transcriptome database established in our laboratory, and two pairs of gene-specific primers (5GSP1, 5GSP2 and 3GSP1, 3GSP2) were designed based on the fragment. Nannochloropsis RNA was extracted with TransZol Plant total RNA extraction kit, reverse transcribed with TransScriptRT reverse transcription kit to form cDNA, and the full-length NgAUREO1 gene was amplified by PCR. The primer sequences are as follows:
[0027] 5GSP1: 5'-GCTTCCTTGTTCAGTTCCTTCG-3'
[0028] 5GSP2: 5'-CAACCACTACCTCCGCCTCA-3'
[0029] 3GSP1: 5'-TACATCTGGCACTACGGGGC-3'
[0030] 3GSP2: 5'-GCTAACACCGACGCTCAAACT-3'.
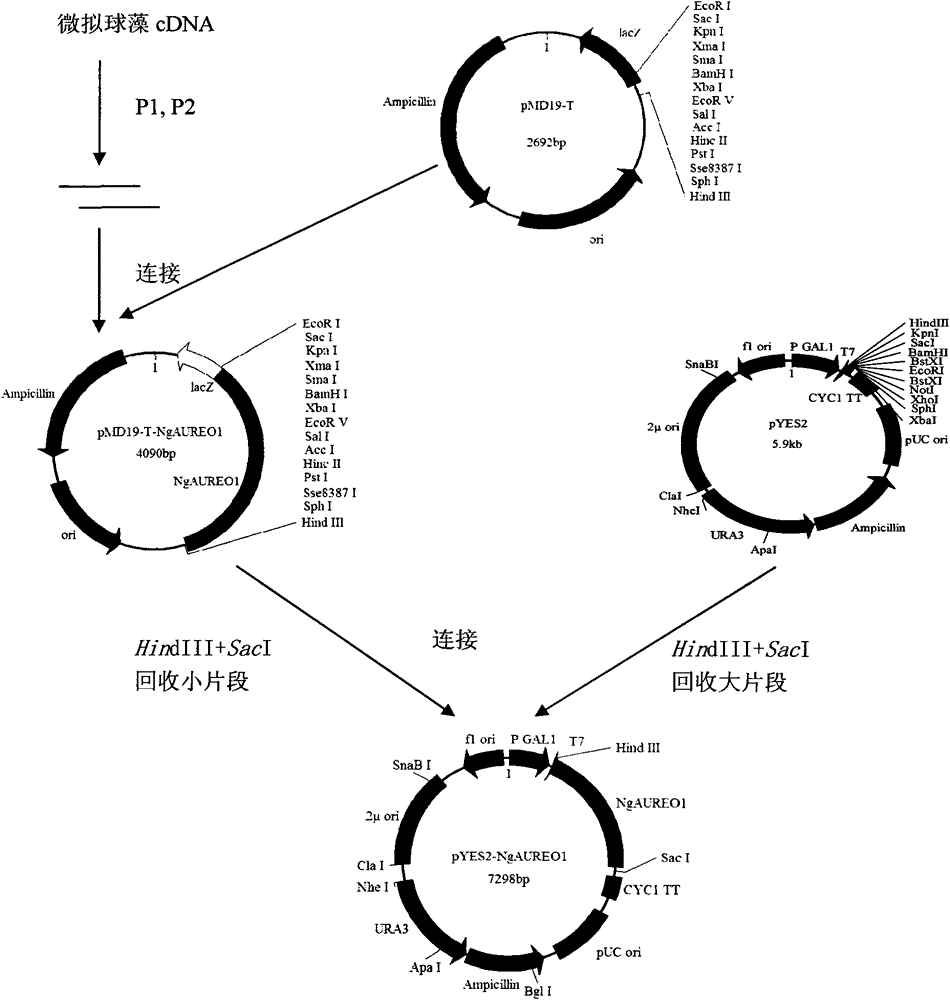
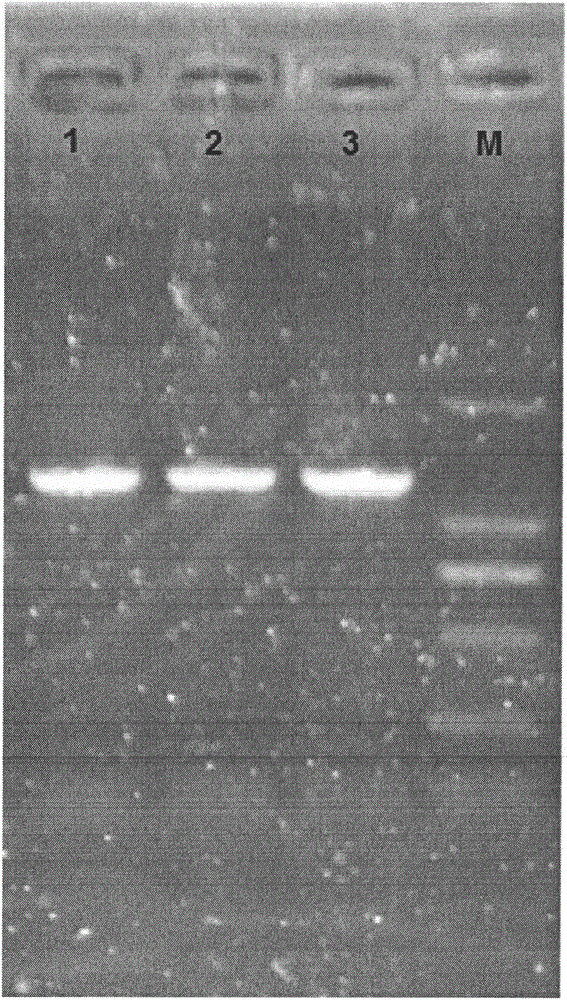
[0031] The NgAUREO1 product amplified by PCR was purified and recovered, connected to the pMD19-T vector, and sent to Shanghai Sangong for sequence determination. Sequencing results showed that the ORF of the gene was 1398bp; the 5' non-coding region was 192bp; the 3' non-codi...
Embodiment 2
[0079] Example 2 Construction of yeast expression vector pYES2-NgAUREO1:
[0080] The NgAUREO1 gene was connected to the pMD19-T vector and named pT-NgAUREO1. Digest pT-NgAUREO1 with HindlII and SacI to recover small fragments, and at the same time digest pYES2 with HindlII and SacI to recover large fragments, connect the small fragments and large fragments, transform Escherichia coli competent cells and carry out PCR and enzyme digestion identification Finally, pYES2-NgAUREO1 was obtained.
[0081] During the operation, plasmid extraction, enzyme digestion, recovery, connection, cell transformation, enzyme digestion identification and PCR identification of recombinant plasmids were all carried out according to the steps described in Example 1.
Embodiment 3
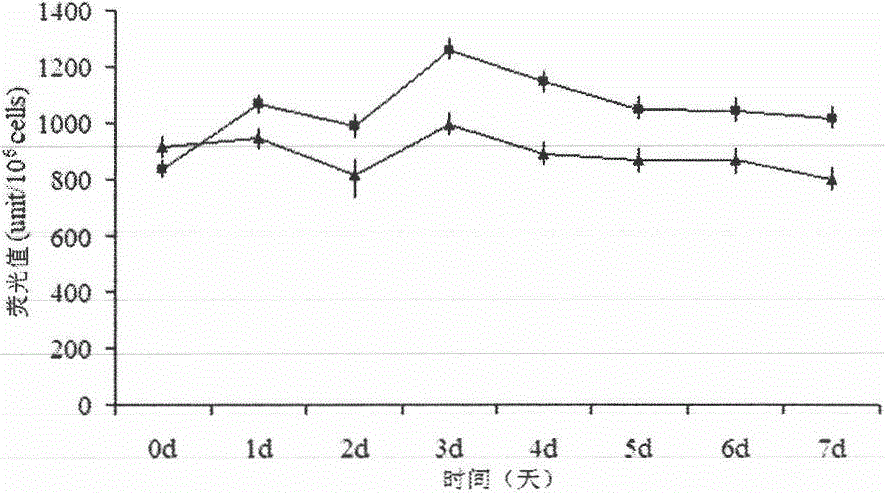
[0082] Genetic Transformation of Example 3 Saccharomyces cerevisiae:
[0083] Preparation of yeast competent cells:
[0084] (1) Inoculate Saccharomyces cerevisiae into 100ml of YPD medium overnight at 30°C, 250rpm, until the density reaches 1×108cells / ml;
[0085] (2) Place the cells on an ice bath for 15 minutes to stop growth;
[0086] (3) Divide the cells into two sterilized 250ml centrifuge tubes, and centrifuge at 3000rpm at 4°C for 5 minutes;
[0087] (4) Remove the supernatant, and place the centrifuge tube on ice;
[0088] (5) Add 50ml of sterilized ice-cold water, vortex to resuspend, set the volume to 50ml in each centrifuge tube, and centrifuge at 3000rpm at 4°C for 5 minutes;
[0089] (6) Repeat operation according to (5) with 25ml of sterilized ice-cold water;
[0090] (7) Add 4ml of sterilized ice-cold 1M sorbitol to resuspend the cells and transfer to an iced 30ml centrifuge tube, centrifuge at 3000rpm at 4°C for 5 minutes, discard the supernatant;
[0091...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com