Trachinotus ovatus peroxiredoxin gene
The technology of peroxidase and reductase of pomfret and trevally is applied in the field of recombinant protein, which can solve the problems of restricting sustainable development, research of oval pomfret treval peroxidase reductase gene, high mortality rate and the like, and achieves good reducing activity and reduction. effect of effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Acquisition of the cDNA gene of peroxide reductase Prx1 in pomfret ovata
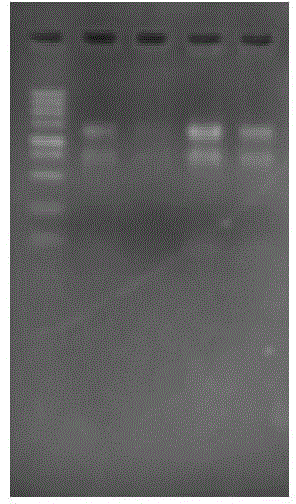
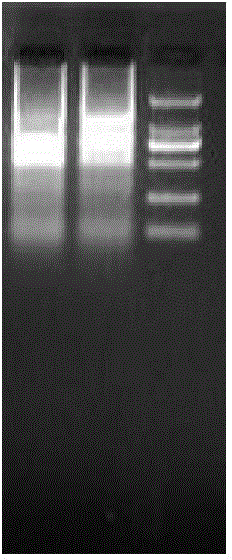
[0040] The full-length gene of peroxide reductase Prx1 of pomfret ovata of the present invention is obtained by sequencing the transcriptome, and the cDNA sequence of peroxide reductase of pomfret ovata is specifically shown in SEQ ID NO.1. Liver total RNA of pompano ovata ( figure 1 Shown) The cDNA obtained by reverse transcription according to the instructions of M-MLV reverse transcriptase was used as a template (see figure 2 ). Reverse transcription reaction conditions: incubate at 70°C for 5 minutes, ice-bath for 5 minutes, add M-MLV reverse transcriptase master mix, incubate at 42°C for 90 minutes, inactivate at 70°C for 15 minutes.
[0041] The amino acid sequence of peroxide reductase Prx1 in pomfret ovata was predicted by software DNAStar7.1. Specifically, it is shown in SEQ ID NO.2.
Embodiment 2
[0042] Example 2 Synthesis of Oval Pomfret Peroxide Reductase Prx1 Gene
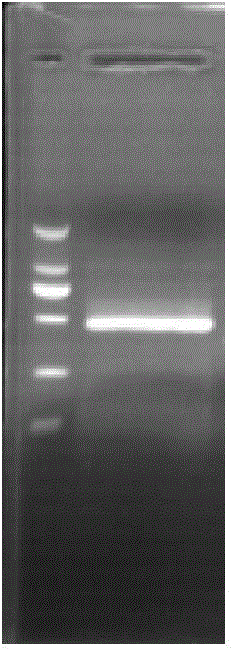
[0043]According to the complete cDNA sequence of the peroxide reductase Prx1 gene of the pomfret pomfret in Example 1, a pair of primers were designed and synthesized, and the upstream primer was a restriction enzyme cut with BamH I before 21 bases from the 128th base site and 3 protective bases (5'-GGTGGATCCATGTCTGCTGGAAATGCTAAG-3'), a total of 30bp; the downstream primer is the first 21 bases from the 701st base plus HindⅢ restriction enzyme site has been 2 Protected base (5'-CGAAGCTTTTACTGCTTGGAGAAGAACTC-3'), 29bp in total. Using the cDNA library of the peroxide reductase Prx1 of the pomfret ovata as a template, the DNA sequence corresponding to the amino acid sequence from the 1st to the 197th amino acid sequence of the peroxide reductase of the pomfret ovata was amplified by PCR method, which is the pompano ovata The corresponding sequence of the mature peptide of trevally peroxide reductase Prx1, ...
Embodiment 3
[0045] Example 3 Construction of an Escherichia coli expression vector containing the peroxide reductase gene of pomfret ovata
[0046] After the PCR product amplified in the above-mentioned embodiment 2 was digested by BamH I and Hind III, the digested product was recovered with the PCR product purification kit of AxyGen Company, and the oval pomfret peroxide reductase gene fragment of about 590bp was isolated and purified; expression The vector pRSET A was also digested with BamH I and HindⅢ, and the digested product was subjected to agarose gel electrophoresis to separate and purify a large fragment of 2.9 kb. Mix, connect with T4 ligase overnight at 16°C and transfer to Escherichia coli BL21 with standard calcium chloride transformation method, screen for ampicillin-resistant transformants, extract plasmids with standard methods, and screen for recombinants with a size of about 3.5 kb. The recombinant mass was double-digested with restriction endonucleases BamH I and HindⅢ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com