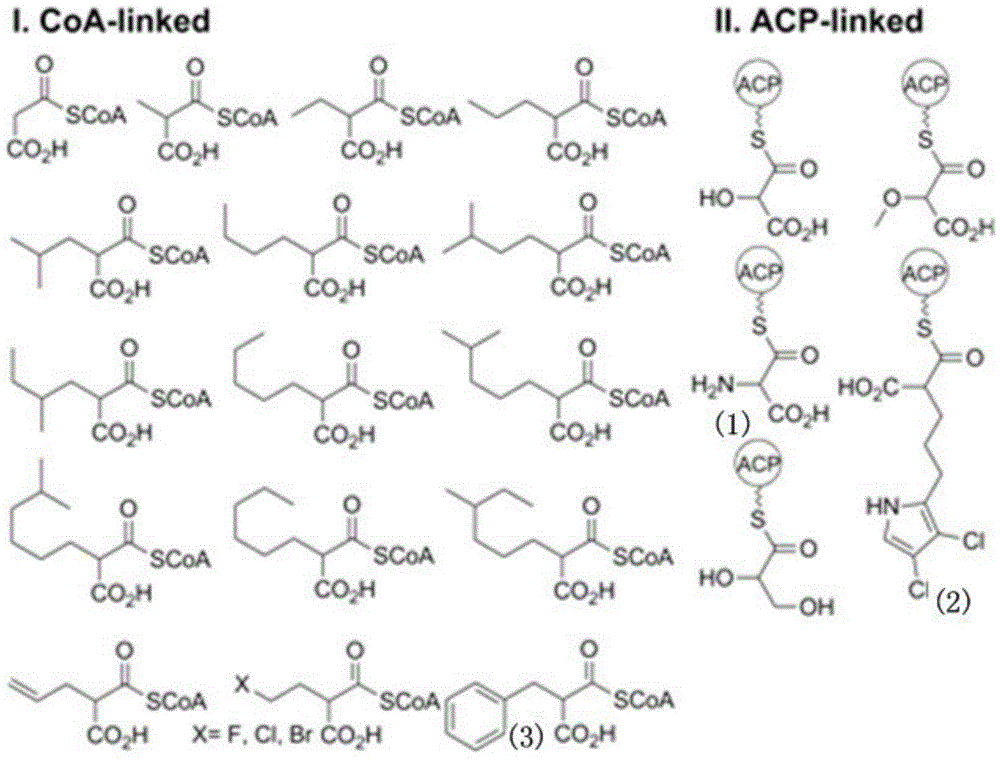
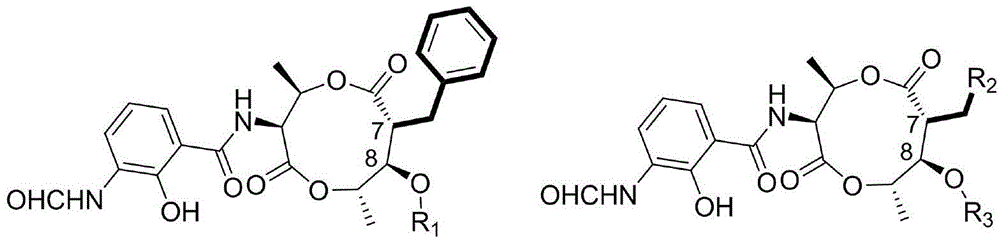
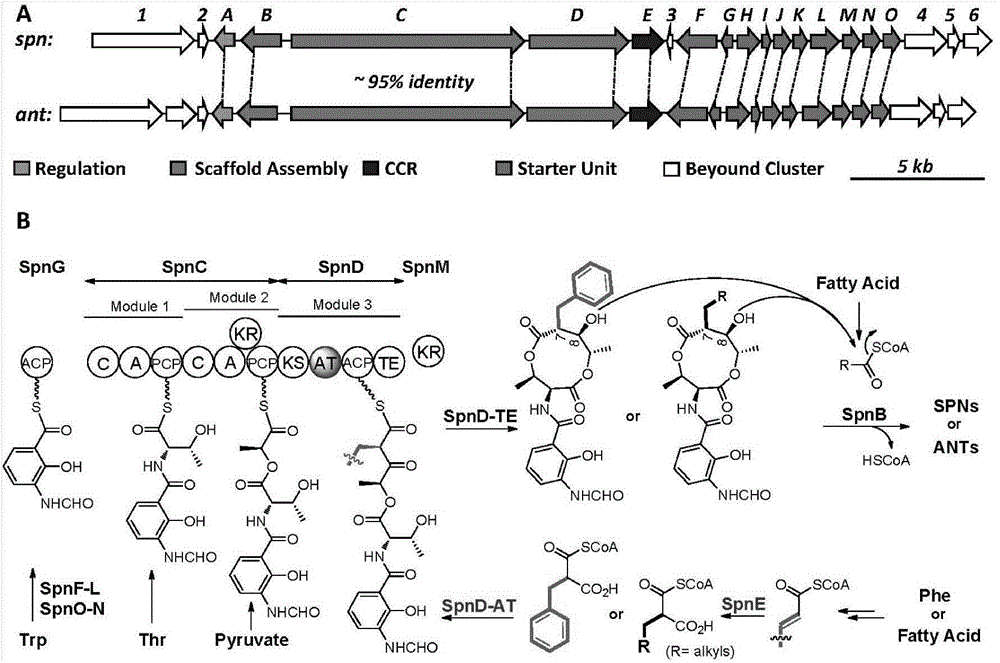
Biosynthesis method for side chain introducing amino acid source to polyketone skeleton and related genes
A technology of biosynthesis and polyketide skeleton, applied in biochemical equipment and methods, plant genetic improvement, botany equipment and methods, etc., can solve the problems of high preparation cost, low incorporation efficiency, limitations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] [Example 1] splenocin producing bacteria S.sp .CNQ431 genetic operating system establishment
[0072] 1. Donor Bacteria Preparation
[0073] Transform the plasmid containing the target into E. coli ET12567, pick a single colony on the plate and inoculate it into 5mL LB (containing the corresponding antibiotic) for overnight culture, pipette 4 mL of the bacterial solution into 50 mL LB (containing the corresponding antibiotic), place it in a shaker at 37 ℃, 250 rpm Cultivate until OD600=0.4 - 0.6. After centrifugation at 6000 rpm for 4 min, the bacteria were washed twice with 20 mL LB to remove antibiotics, and then resuspended in 1 mL LB as DNA donor bacteria.
[0074] 2. Recipient Bacteria Preparation
[0075] Take out a tube of spore suspension (6.7x109 cells / mL) stored at -80°C in 20% glycerol, centrifuge at 8000 rpm for 3 min to remove glycerol, and wash the spores twice with 0.5 mL TES buffer (0.05 M, pH 8.0) to Glycerol was removed, then resuspended with 50...
Embodiment 2
[0078] [Example 2] S.sp . Construction and screening of CNQ431 gene library
[0079] 1. Mini-digestion experiment
[0080] Add 10 u / μL of Alu ⅠRestriction endonuclease (Takara) was diluted with 10 × L buffer and sterilized water to make the working concentration (1×) of the buffer, and 15 μL of the enzyme concentration of 0.33 U / μL, and then prepare 250 μl of the reaction system (containing CNQ 431 gDNA 40 μl, 10× L buffer 25 μl), then divide the 250 μl reaction system into 1 × 50 μl and 7 × 25 μl, then take 2 μl and dilute to 0.33 u / μl in advance Alu Ⅰ Add 50 μl of the reaction system in the first tube, mix well and transfer 25 μl to the second tube of 25 μl, repeat this transfer step 7, all the above steps must be performed on ice, and then all the systems 37 After digestion at ℃ for 15 min, inactivation at 70 °C for 10 min, electrophoresis at 40 V, 0.5% agarose gel for 12 h in a cold room at 4 °C, staining with ethidium bromide, and observing the quality of enzyme dig...
Embodiment 3
[0093] [Example 3] Construction of mutant or recombinant strains
[0094] 1. SpnC gene disruption
[0095] Primer 5'-CG GAATTC GAGCGGCTGACCTCGTTCTG-3' and 5'-C CAAGCTT G
[0096] ACGACCGTGCACAGTCCGT-3'( EcoR I and Hind III restriction site is underlined) using the genome of CNQ431 as a template, a 3.08 kb middle fragment of the spnC gene was amplified and connected to PMD19-T, which was confirmed to be correct by sequencing. Bam HI- Hind III digestion, because the middle fragment of spnC has a Bam HI restriction site, so the excised fragment is only 1.1 kb, and then ligated into the plasmid pYH7 Bgl II- Hind III site. The constructed plasmid was named pWHU2406, and then transferred to CNQ431 through the genetic operating system. The grown binders were streaked onto MS plates to induce gene recombination. Pick the colony from the MS plate and inoculate it on the MS plate containing 50 μg / mL apramycin, culture at 30°C, and screen out Δ spnC The mutant stra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com