Molecular identification method used for siniperca chuatsi, siniperca scherzeri and hybrid f1 of siniperca chuatsi and siniperca scherzeri
A technology of molecular identification and cocked mandarin fish, which is applied in the determination/inspection of microbes, biochemical equipment and methods, etc., can solve the problems of mixed germplasm and difficult identification, and achieve moderate price, reduced detection time, and wide applicability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] A method for molecular identification of mandarin mandarin fish, mandarin mandarin fish and their hybrid offspring, the steps are:
[0030] 1. Genomic DNA extraction:
[0031] Cut about 0.5 mg of fin ray tissue from each fish, and extract genomic DNA according to the method and steps shown in the blood, cell, tissue genomic DNA extraction kit (spin column type) produced by Tiangen Biochemical Technology (Beijing) Co., Ltd., and use The concentration of DNA was detected by ultraviolet spectrophotometer, and the purity of DNA was detected by agarose gel with a concentration of 1.5%.
[0032] 2. Amplification of target DNA fragments:
[0033] That is, PCR is performed.
[0034] 1) The sequence of a pair of microsatellite primers used in PCR reaction is:
[0035] Primer forward sequence: 5'ATGTAACCGTAAGTGACCTC3',
[0036] Primer reverse sequence: 5'TGTTGTTCAGACGATGACGA3'.
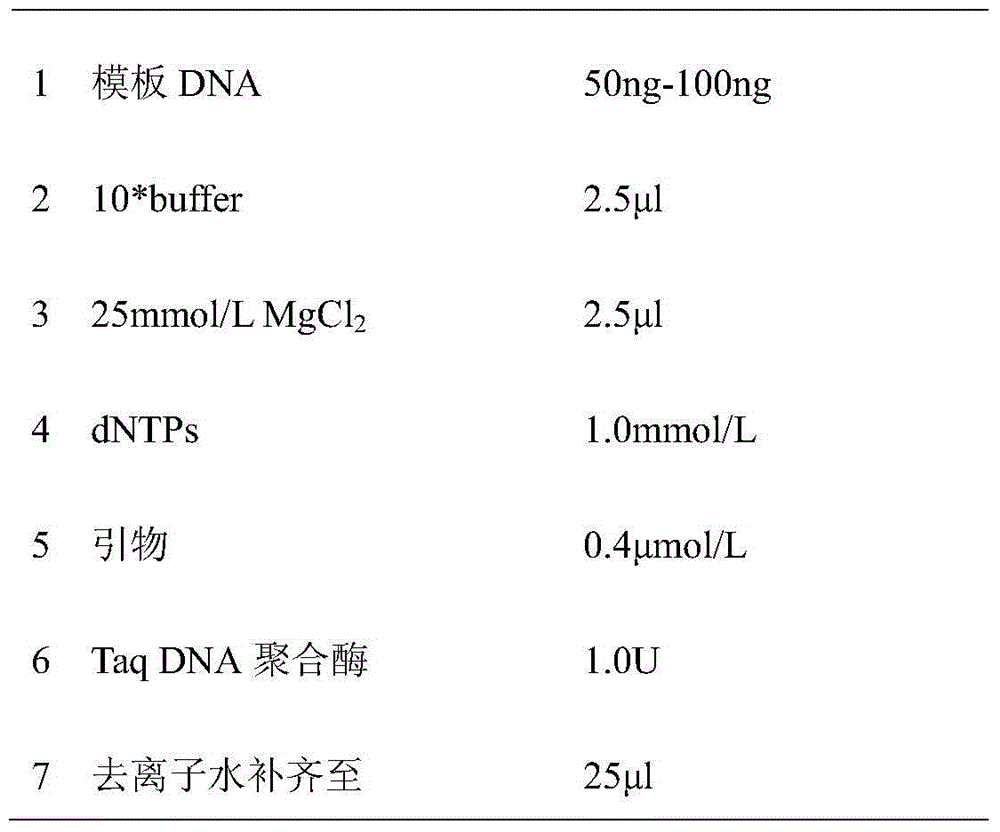
[0037] 2) The PCR reaction system is as follows:
[0038]
[0039]
[0040] Mix and centr...
Embodiment 2
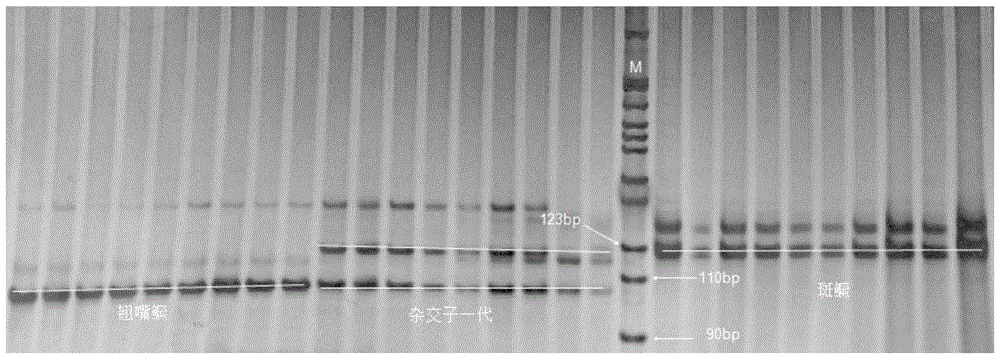
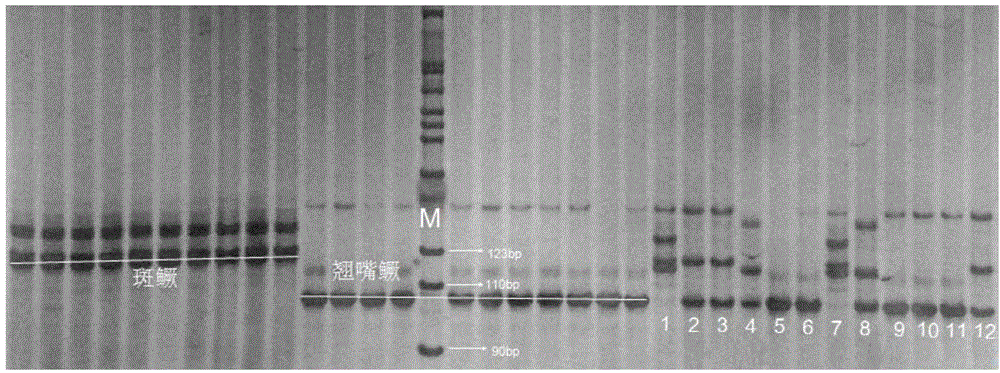
[0049] A verification example of a molecular identification method for Siniperca sinensis, Siniperca sinensis and their hybrid offspring, the steps are:
[0050] 1. Sample source and extraction of genomic DNA:
[0051] 33 mandarin fishes (11 parents of mandarin fish with cocked mouth, 10 parents of mandarin mandarin fish with cocked mouth, 12 offsprings of hybrids of mandarin fish with cocked mouth and mandarin mandarin fish with cocked mouth, 12 offsprings of mandarin mandarin fish with cocked mouth and children of spotted mandarin fish) from Yushun Farming and Animal Husbandry in Qingyuan, Guangdong Fishery Science and Technology Service Co., Ltd. fishery harvested. Cut about 0.5 mg of fin ray tissue from each fish, and extract genomic DNA according to the method and steps shown in the blood, cell, tissue genomic DNA extraction kit (spin column type) produced by Tiangen Biochemical Technology (Beijing) Co., Ltd., and use The concentration of DNA was detected by ultraviolet ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com