Xylose isomerases and their uses
A technology of xylose isomerase and structural domain, applied in the field of XI polypeptide, can solve the problems of no enzyme activity and low enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0120] Materials and methods
[0121] yeast culture
[0122]Unless otherwise stated for a particular example, yeast transformants were grown in SC-ura medium containing about 2% glucose at 30°C for about 24 hours. The medium contains about 20 g of agar, about 134 g of BD Difco TM Amino Acid Free Yeast Nitrogen Base (BD, Franklin Lakes, New Jersey) and about 2 g SC Amino Acid Blend containing about 85 mg of the following amino acid unless otherwise noted (amounts are listed in parentheses): L-Adenine (21.0), L-alanine, L-arginine, L-asparagine, L-aspartic acid, L-cysteine, glutamine, L-glutamic acid, glycine, L- Histidine, Inositol, L-Isoleucine, L-Leucine (173.4), L-Lysine, L-Methionine, P-Aminoic Acid (8.6), L-Phenylalanine , L-proline, L-serine, L-threonine, L-tryptophan, L-tyrosine, L-valine.
[0123] Xylose isomerase activity
[0124] XI activity in cell lysates was assayed using a method based on the method of Kersters-Hilderson et al., 1986, Enzyme Microb. Technol...
Embodiment 2
[0127] Example 2: Activity-Based Discovery Screen of Xylose Isomerase
[0128] Libraries for activity-based discovery ("ABD") screening were in the form of ex vivo phagemids. These libraries were constructed as described in US Patent 6,280,926. The source of these libraries was environmental rumen samples collected from the foregut of dead herbivores.
[0129] E. coli screening strains were constructed to identify genes encoding xylose isomerase activity from environmental libraries. Specifically, the E. coli strain SEL700 was complemented with the pBR322-derived plasmid pJC859 (Kokjohn et al., 1987, J. Bacteriol. 169:1499-1508) containing the E. coli recA gene to generate a wild-type recA expression. type, the SEL700 is recA - , MG1655 derivatives resistant to bacteriophage lambda and containing the F' plasmid.
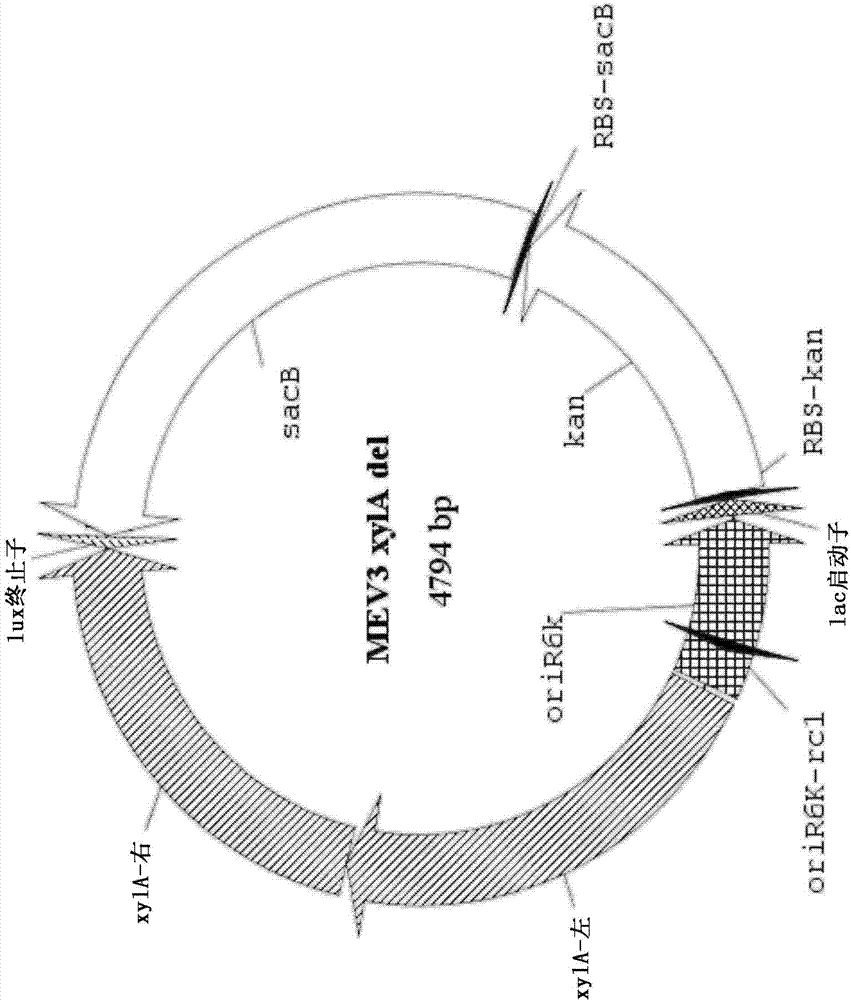
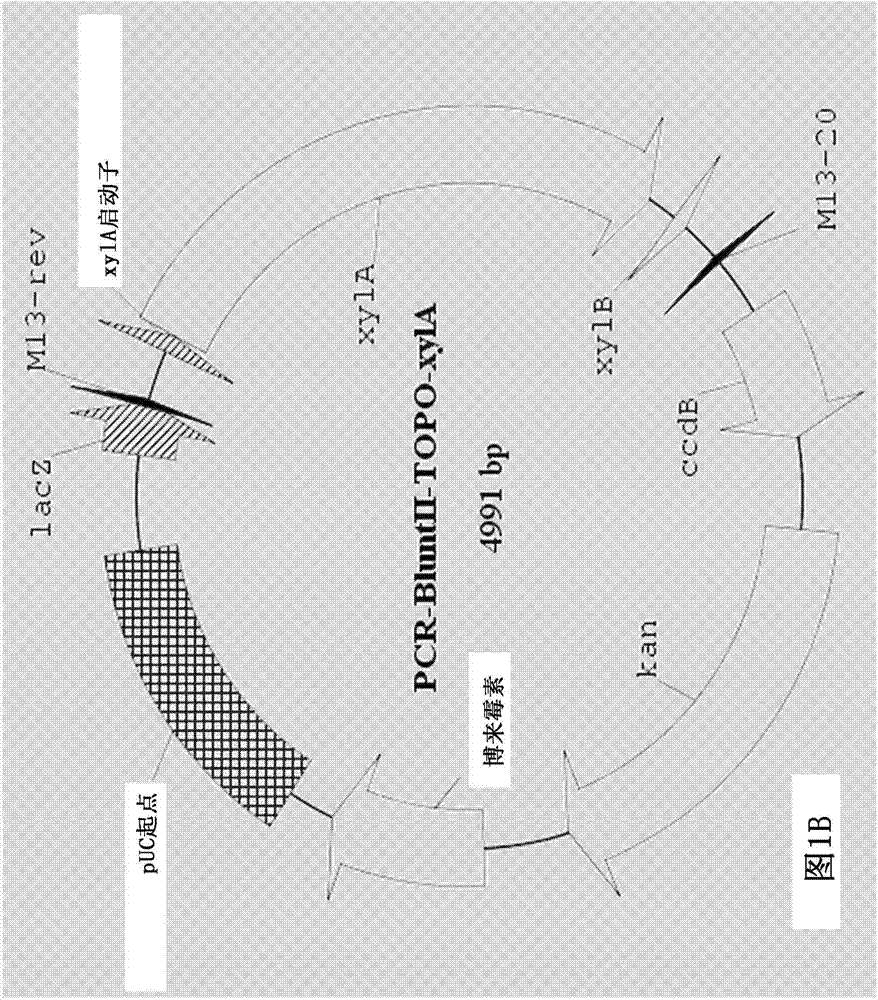
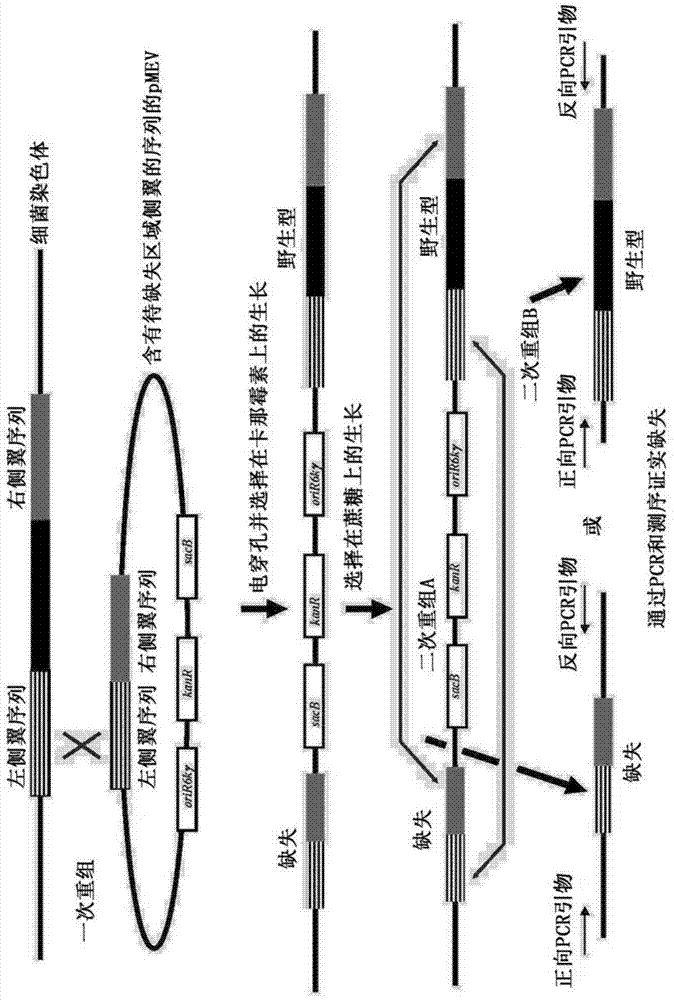
[0130] The entire coding sequence of the endogenous xylA xylose isomerase gene was then deleted using a two-step marker exchange procedure. Briefly, plasmid p...
Embodiment 3
[0138] Embodiment 3: XI sequence analysis
[0139] Plasmids from both the ABD and SBD screens were purified using the ABI3730xl DNA Analyzer and the ABI v3.1 cycle sequencing chemistry for sequencing vector inserts. Identification of putative ORFs was accomplished by identifying the start and stop codons of the longest protein coding region, followed by manual curation based on homology to published xylose isomerase DNA sequences. The identified XI ORFs are listed in Table 2 below, which indicates the sequence and source organism classification for each XI determined from the ABD or SBD library, as well as their assigned sequence identifiers. The putative catalytic domain (based on sequence alignment with other XIs) is underlined.
[0140]
[0141]
[0142]
[0143]
[0144]
[0145]
[0146]
[0147]
[0148]
[0149]
[0150]
[0151]
[0152]
[0153]
[0154]
[0155]
[0156]
[0157]
[0158]
[0159]
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com