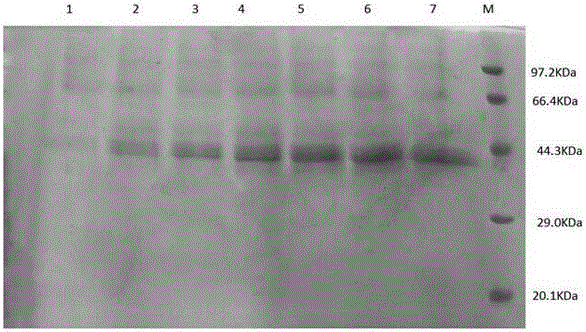
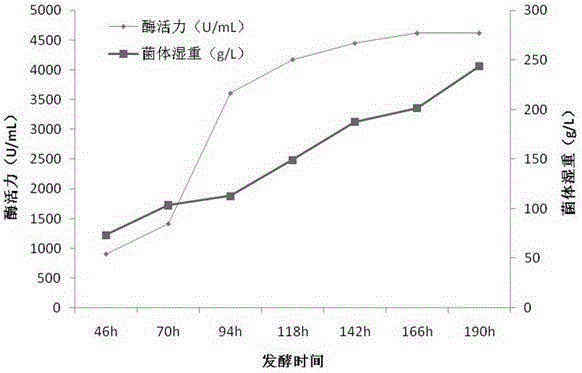
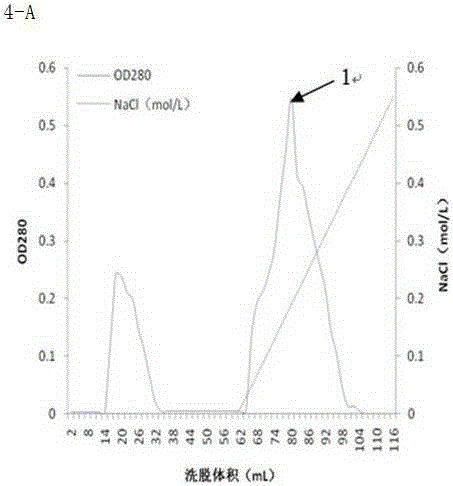
Agarase and preparation method thereof
A technology of agarase and gene, applied in the biological field, can solve the problem of no hydrolysis of carrageenan, and achieve the effect of good decomposition ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The acquisition of embodiment 1 Agarase gene
[0043] Operate according to genomewalkingkit.
[0044] (1) Microbulbiferthermotolerans Separation and extraction of genomic DNA:
[0045] Take 1.5 mL of bacterial culture in a sterilized Ep tube, centrifuge at 12,000 rpm for 1 min, discard the supernatant, and collect the bacterial cells.
[0046] Add 400 μL of lysate (40 mM Tris-acetic acid, 20 mM sodium acetate, 1 mM EDTA, 1% SDS, pH7.8) and mix well, and place in a 37 ° C water bath for 1 h.
[0047] Then add 200μL of 15mol / L sodium chloride solution, mix well and centrifuge at 13000rpm for 15min.
[0048] Take the supernatant, extract twice with phenol and once with chloroform.
[0049] Add twice the volume of absolute ethanol, 1 / 10 volume of potassium acetate (3M, pH8.0), store at -20°C for 1 hour, centrifuge at 13,000 rpm for 15 minutes, discard the supernatant, and wash the precipitate twice with 70% ethanol; place at room temperature After drying, it was dis...
Embodiment 2
[0064] Example 2 Construction of Yeast Recombinant Expression Vector
[0065] With the target gene obtained in embodiment 1, and through to EcoR I and not The pPIC9k plasmid of I double digestion is ligated to obtain the recombinant plasmid pPIC9k-Agarase (such as figure 1 shown).
[0066] Using the obtained pPIC9k recombinant plasmid as a template, PCR was performed with the primer pair composed of primer P1 and primer P2. At the same time, PCR was performed with the primer pair composed of the pMD18-T recombinant plasmid prepared in Example 1 and primer P1 and primer P2. From the DNA Horizontally verify whether the foreign gene insertion is correct. 2 kinds of product sequence lengths that PCR obtains are 1311bp, and in embodiment 1 from Microbulbiferthermotolerans The sequence and other characteristics of the Agarase original gene obtained in the above method are consistent, so it can be known that the insertion site, direction and sequence of the target gene are ...
Embodiment 3
[0067] Example 3 Pichia pastoris fermentation production recombination Agarase
[0068] The recombinant plasmid prepared in Example 2 was subjected to Sac I digested to obtain the linearized plasmid pPIC9k-Agarase1.
[0069]Take 50 μg of the constructed linear recombinant plasmid DNA and directly add it to competent cells (Pichia pastoris GS115) that are still below 0°C; add 1.0 ml of solution II containing 5 μg / ml salmon sperm DNA (40% (w / v ) polyethylene glycol 1000, 0.2MN, N-dihydroxyethylglycine, pH8.35), or first add 1.0ml of solution II, then add 5μL of 1mg / ml salmon sperm DNA and try to mix the two completely ; keep warm in a water bath at 30°C for more than 1 hour, and mix gently every 15 minutes; keep warm at 42°C for 10 minutes; Hydroxyethylglycine, pH8.35) to resuspend the cells; centrifuge at 3000×g for 5 min at room temperature, remove 800 μL supernatant, and use the remaining 200ul supernatant to resuspend the cells; spread 200 μL of the cells on a YPD plate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com