Method and special kit for detection of porcine cytomegalovirus antibody
A kit, antigen protein technology, applied in the direction of viruses/phages, viruses, viral peptides, etc., can solve the problems of limited detection sensitivity, inability to perform high-throughput testing, and time-consuming serology and fluorescent antibody detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
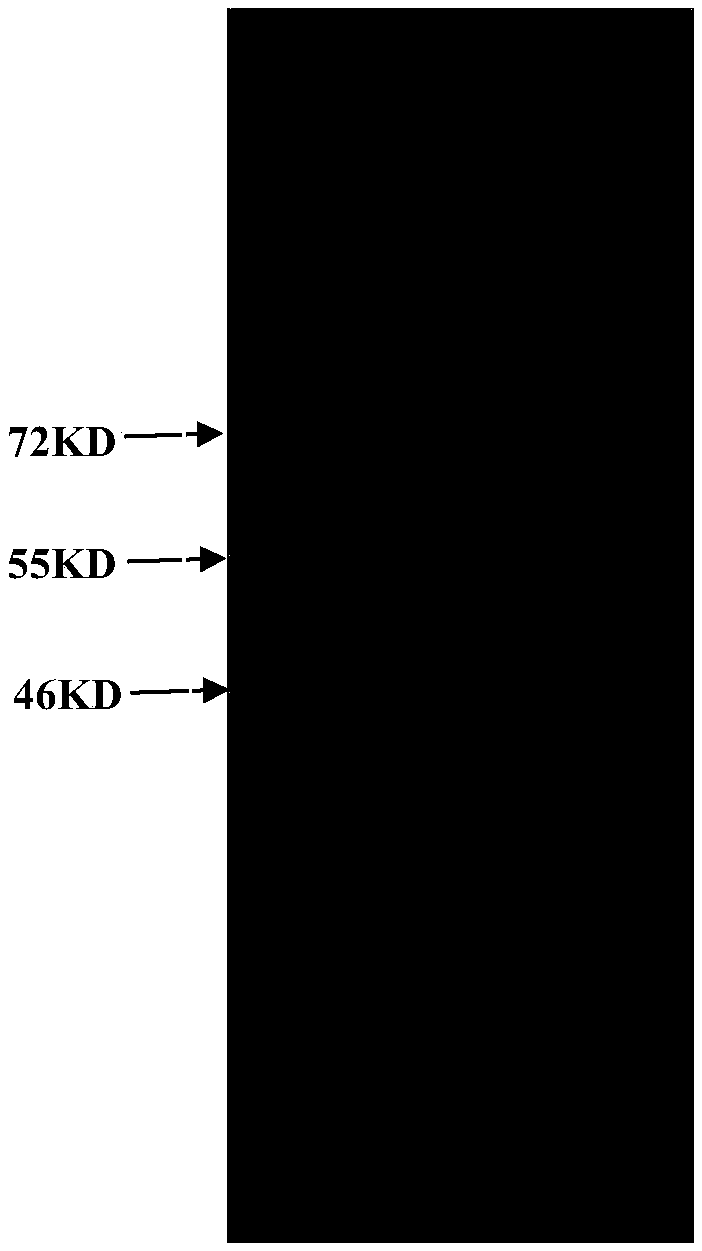
[0088] Embodiment 1, the preparation of antigenic protein
[0089] 1. Carrier Construction
[0090] The double-stranded DNA molecule shown in sequence 1 of the artificially synthesized sequence listing was inserted between the BamHI recognition sequence and the HindIII recognition sequence of plasmid pET-32a(+), to obtain recombinant plasmid pET-32a-K.
[0091] The double-stranded DNA molecule shown in sequence 1 of the sequence listing encodes the protein shown in sequence 2 of the sequence listing. The protein shown in Sequence 2 in the sequence listing is the antigenic protein.
[0092] In the recombinant plasmid pET-32a-K, the double-stranded DNA molecule shown in sequence 1 of the sequence listing is fused with part of the nucleotides on the vector backbone to form a fusion gene shown in sequence 4 of the sequence listing, expressing sequence 3 of the sequence listing Antigenic proteins with His tags are indicated.
[0093] 2. Preparation of antigenic protein
[0094]...
Embodiment 2
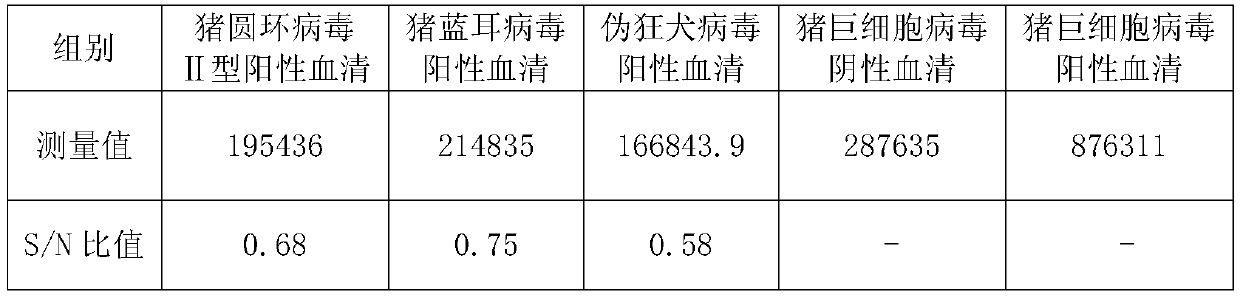
[0101] Embodiment 2, establishment of the method for detecting porcine cytomegalovirus antibody in the serum to be tested
[0102] The method for detecting the porcine cytomegalovirus antibody in the serum to be tested is as follows:
[0103] 1. Take the microtiter plate and coat it with the antigenic protein prepared in Example 1;
[0104] 2. Add the serum to be tested or porcine cytomegalovirus negative serum and incubate;
[0105] 3. Add HRP-labeled goat anti-pig IgG antibody and incubate;
[0106] 4. Add luminescence solution and incubate;
[0107] 5. Detection by chemiluminescence detector.
[0108] Then judge as follows: if the absorbance value of the serum to be tested is greater than 2.1 with the absorbance value of the porcine cytomegalovirus negative serum, the serum to be tested contains porcine cytomegalovirus antibody; The absorbance ratio is below 2.1, and the serum to be tested does not contain porcine cytomegalovirus antibody.
Embodiment 3
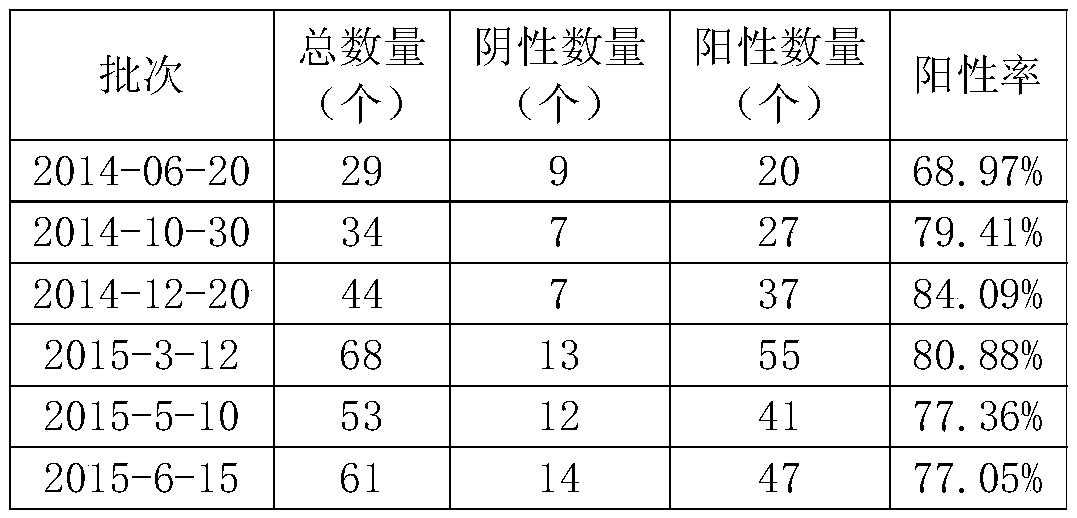
[0109] Embodiment 3, the optimization of the optimal detection condition of the method for detecting the porcine cytomegalovirus antibody in the serum to be tested
[0110] In order to save reagents and workload, the single factor variable was used to determine the optimal value, and then the optimal detection conditions were determined.
[0111] 1. Determination of antigenic protein coating concentration
[0112] The steps to determine the coating concentration of antigenic protein are as follows:
[0113] 1. Take the microtiter plate, and use the antigenic protein prepared in Example 1 as the coating source to coat (dilute the antigenic protein solution prepared in Example 1 with the coating liquid, and set the following coating concentrations respectively: 3.050 μg / ml, 1.525 μg / ml, 0.610μg / ml, 0.305μg / ml, 0.203μg / ml, 0.153μg / ml, 0.122μg / ml, 0.102μg / ml, add 100μl to each well), seal with parafilm, and coat at 4°C 20h, then discard the supernatant, wash with washing solutio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com