Kluyveromyces marxianus-derived alkyl peroxide reductase and thioredoxin reductase and application thereof
A technology of alkyl peroxide and thioredoxin, applied in the application of improving the stress tolerance of Saccharomyces cerevisiae, in the field of gene and amino acid sequence of thioredoxin reductase KmTrxR, can solve limited problems and less functional research And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 Construction of Saccharomyces cerevisiae strains overexpressing alkyl peroxide reductase KmTPX1
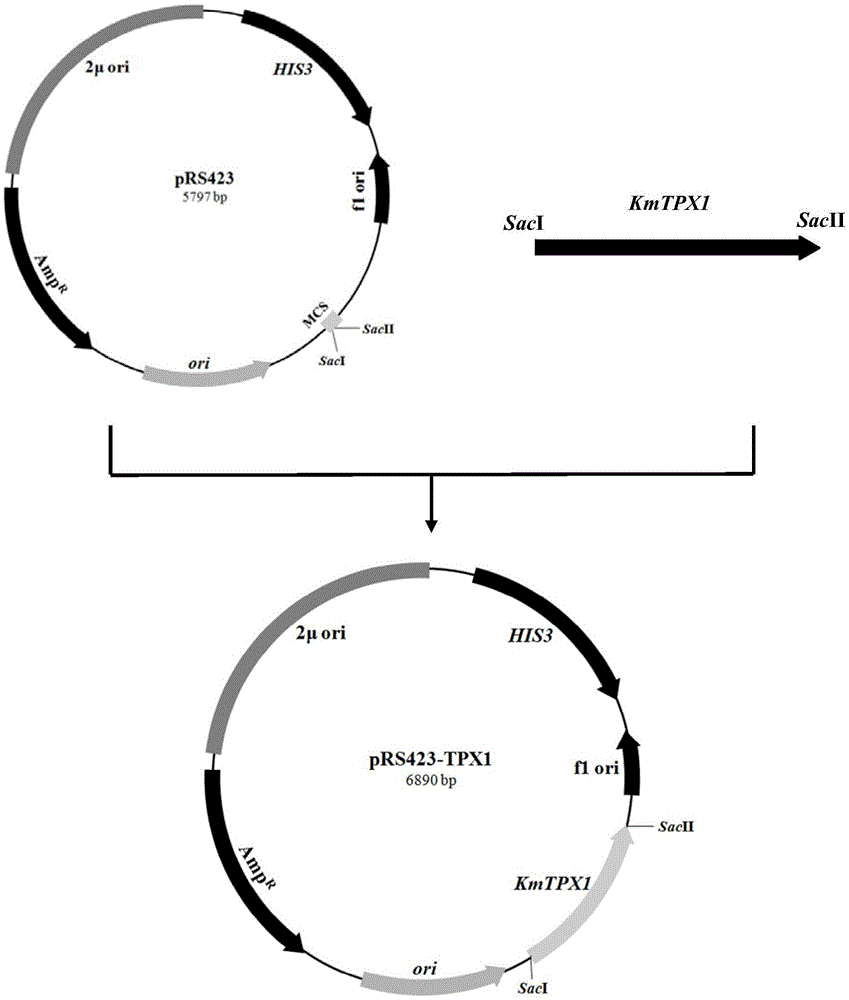
[0077] 1. Cloning of KmTPX1 gene
[0078] PrimerPremier5.0 primer design software was used to design the upstream primer TPX1-F, 5'-CTTGAGCTCAATGTCTCGTCTCGTCTCGT-3' and downstream primer TPX1 based on the complete sequence of the alkyl peroxide reductase KmTPX1 gene of K.marxianus (obtained by transcriptome sequencing) -R, 5'-TCCCCGCGGGGCTAAGCCAATAACTTATT-3', introducing a SacI restriction site at the 5' end and a SacII restriction site at the 3' end, respectively. The K. cicerisporus CBS4857 genome was used as a template for PCR amplification reaction. Among them, the PCR reaction system (50μL):
[0079]
[0080] PCR reaction conditions:
[0081]
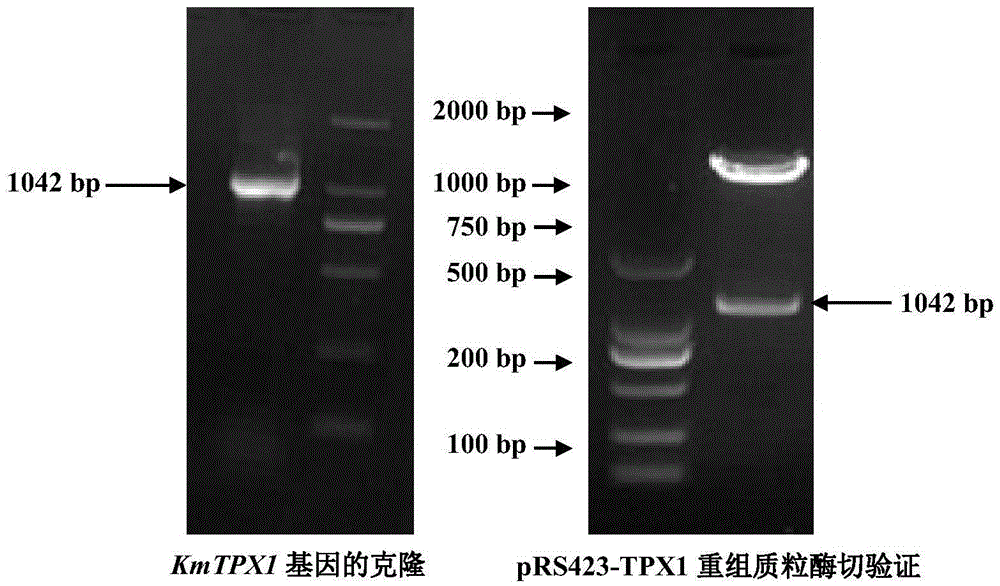
[0082] After the PCR reaction, 2 μL of the PCR product was taken for agarose gel electrophoresis detection, and the product contained a total of 1093 nucleotides, including a 478 bp promoter and a 594 bp codin...
Embodiment 2
[0115] Example 2 Construction of a Saccharomyces cerevisiae strain overexpressing thioredoxin reductase KmTrxR
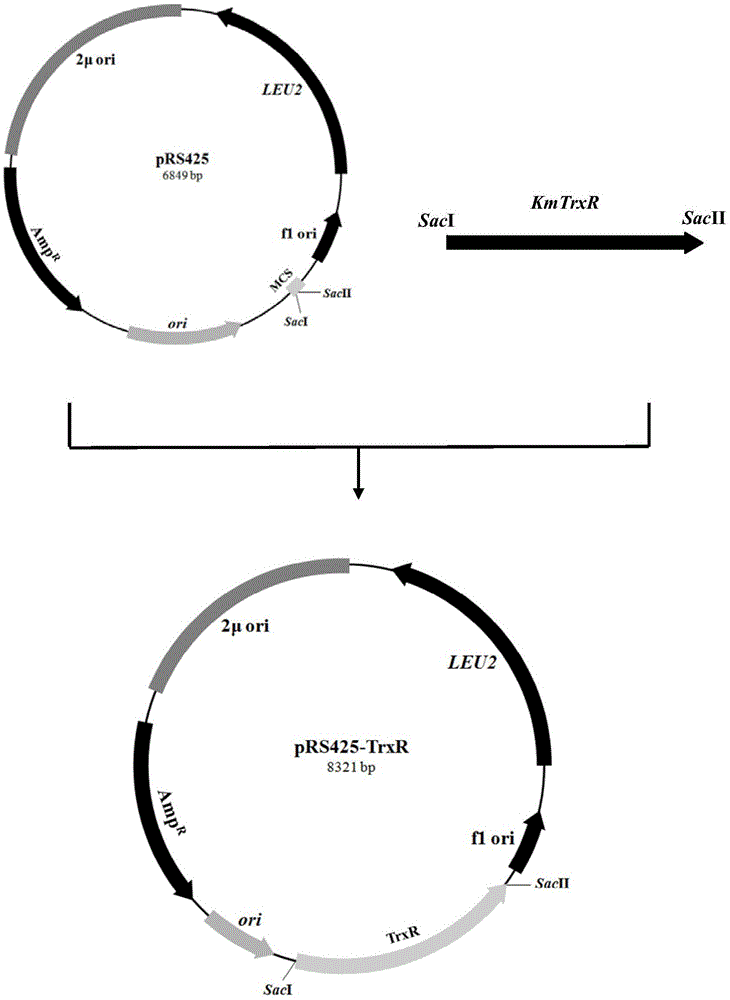
[0116] Similarly, using PrimerPremier5.0 primer design software, based on the complete sequence of K.marxianus thioredoxin reductase KmTrxR (acquired by transcriptome sequencing), the upstream primer TrxR-F, 5'-TCCGAGCTCCCCATGTCAAATGATGAAACG-3' and downstream primer TrxR were designed -R, 5'-TCCCCGCGGACGAGGAACCAACCTTTATT-3', introducing a SacI restriction site at the 5' end and a SacII restriction site at the 3' end, respectively. The K. cicerisporus CBS4857 genome was used as a template for PCR amplification reaction. The PCR reaction system and reaction conditions were the same as in Example 1, and the obtained PCR product contained a total of 1472 nucleotides, including its own promoter of 497 bp and a coding sequence of 960 bp.
[0117] The purified PCR product was combined with Saccharomyces cerevisiae multi-copy vector pRS425 ( figure 2 ) for ligation react...
Embodiment 3
[0119] Embodiment 3 Real-time quantitative PCR verifies the expression intensity of KmTPX1 gene and KmTrxR gene
[0120] In order to confirm that the recombinant plasmids containing KmTPX1 gene and KmTrxR gene constructed in Examples 1 and 2 can be successfully transcribed in Saccharomyces cerevisiae, we used real-time quantitative PCR to verify the expression intensity of these two genes.
[0121] First, the expression of the KmTPX1 gene was verified. The overexpressed KmTPX1 Saccharomyces cerevisiae strain (named TPX1) and the control yeast strain (named 423) containing the empty plasmid pRS423 were both cultured in SD medium lacking histidine (0.67% yeast nitrogen base, 2% glucose and other essential amino acid nutrients, SD-His). The two activated yeasts were respectively inoculated into SD-His medium according to the inoculum amount of 1% (v / v), and cultured at 30° C. and 150 rpm until the logarithmic growth phase (16-18 h). Cells were collected for total RNA extraction...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com