A method for pretreatment of yeast β-glucan in milk or dairy products
A dairy product and glucan technology, applied in the field of pretreatment, can solve problems such as lack of detection standards and related technologies, complex dairy matrix, and influence on the extraction and purification of yeast β-glucan
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] (1) Proteolysis: Add 0.1% (v / v) to 3 samples of yeast β-glucan milk neutral protease stock solution and Equal volume mixture of alkaline protease stock solution, enzymatic hydrolysis at 45°C for 2 hours. The 0.1% (v / v) of neutral protease stock solution and The equal-volume mixed solution of the alkaline protease stock solution corresponds to the enzyme activity of the neutral protease in the milk to be enzymolyzed as 5.0×10 -4 AU / mL, the enzyme activity of the alkaline protease is 1.4×10 -3 AU / mL.
[0080] (2) Filter filtration separation: use 0.8 μm filter paper, 0.8 μm double-layer filter paper and 0.7 μm / 0.8 μm double-layer filter paper (corresponding to samples 1a-1c) to filter the enzymatic solution obtained in step (1);
[0081] (3) Acid hydrolysis: add 1 mol / L hydrochloric acid to the retentate obtained in step (2), and hydrolyze at 121° C. for 1 hour.
[0082] The results showed that when using 0.8 μm filter paper, the retentate appeared on the filter...
Embodiment 2
[0084] Adopt the same step (1) and step (3) as in Example 1.
[0085] Before implementing step (2), dilute the protein hydrolyzate obtained in step (1) by 1 time with 80° C. NaOH aqueous solution. Subsequently, use 1.6 μm, 1.0 μm and 0.7 μm glass fiber filter paper (corresponding to samples 2a-2c) to filter the precipitate obtained in step (1), and use pure water or deionized water to carry out negative pressure pumping on the retentate Filter and wash 2-3 times.
[0086]The results showed that the filter paper with smaller pore size retained more dextran, but its filtration speed was significantly lower than that of filter paper with large pore size. In addition, when the temperature of the milk sample drops, protein and fat deposits tend to occur, resulting in blockage of the filter pores. It was determined that the dextran contents of samples 2a-2c were 31.mg / 100g, 38.9mg / 100g and 45.9mg / 100g respectively.
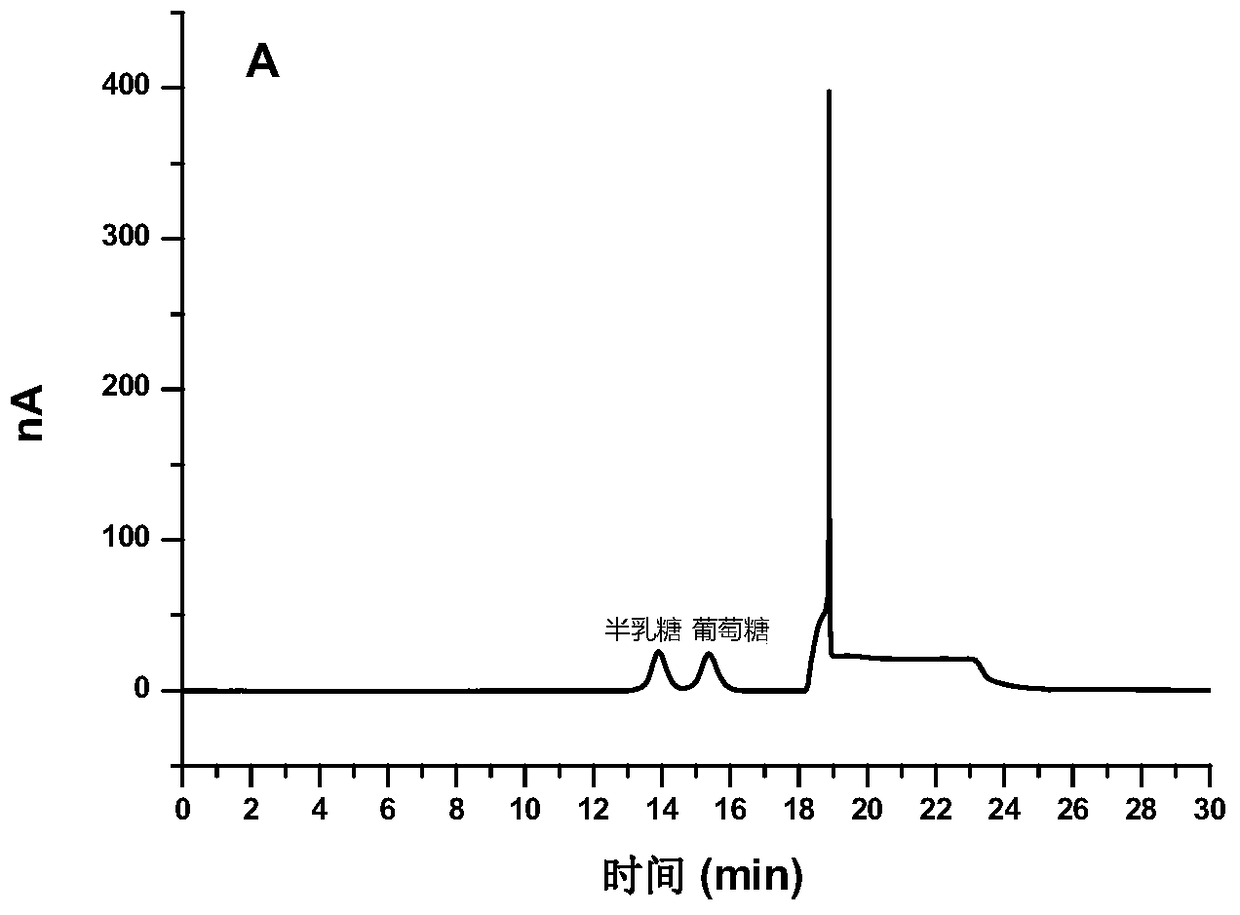
[0087] Such as Figure 3C As shown, the milk sample treated by...
Embodiment 3
[0089] Embodiment 3: Sensitivity, accuracy and precision analysis of analysis step
[0090] Use pure water to prepare glucose standard solutions and sample solutions with a series concentration of 0.5, 1.0, 4.0, 8.0, 10.0, 15.0, and 20.0 μg / mL, and evaluate the sensitivity, accuracy, and precision of the analysis.
[0091] Under the same chromatographic conditions as in Example 2, the linear regression equation obtained by measuring the glucose standard solution is Y=2.865X+0.395, the linear range is 0.5~20 μg / mL, the linear correlation coefficient is 0.9995, and the detection limit is 0.1 μg / mL. The relative standard deviation was 0.15% (n=6).
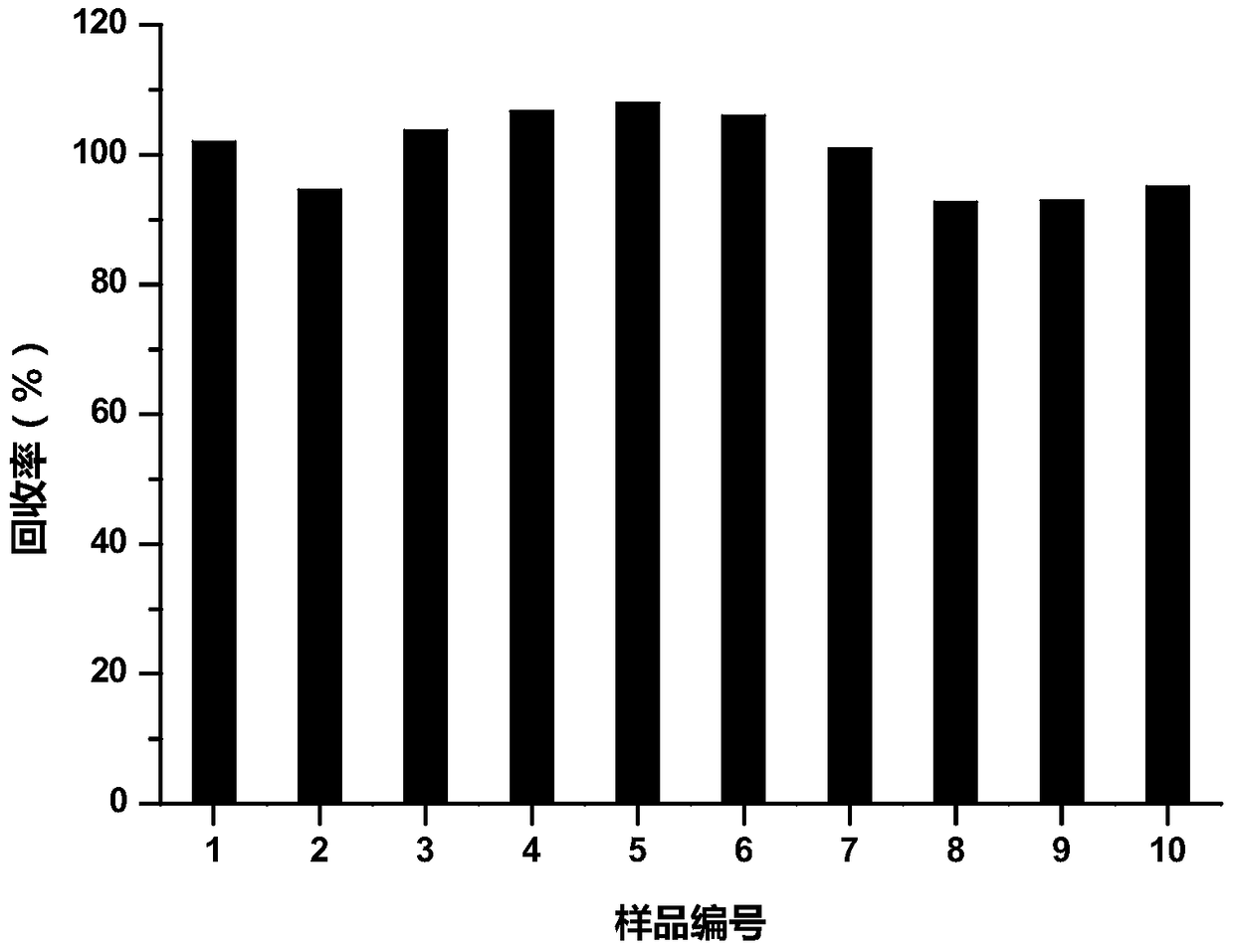
[0092] Taking the concentration of yeast β-glucan added in dairy products at 42.00mg / 100g as the standard sample, prepare samples with three addition levels of 70%, 100% and 130% of the standard sample, and determine the sample recovery rate at the three concentrations : when the addition amount was 70%, the average measured value wa...
PUM
| Property | Measurement | Unit |
|---|---|---|
| degree of polymerization | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com